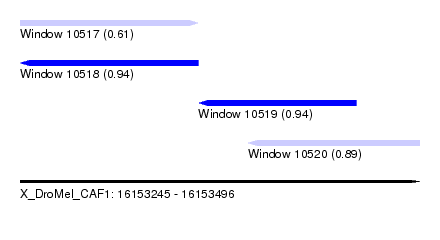
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 16,153,245 – 16,153,496 |
| Length | 251 |
| Max. P | 0.940185 |

| Location | 16,153,245 – 16,153,357 |
|---|---|
| Length | 112 |
| Sequences | 3 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 89.58 |
| Mean single sequence MFE | -26.76 |
| Consensus MFE | -23.12 |
| Energy contribution | -23.46 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.79 |
| Structure conservation index | 0.86 |
| SVM decision value | 0.15 |
| SVM RNA-class probability | 0.606908 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16153245 112 + 22224390 GCAAACGGGUUUACAGAACUCGCGCAUUCACACCCGAAUAUCUCUGUGUGUACACUGUACAUCUGCGCGUACAUUACCCGCAUGCACACAUGUACAUAUGCAAGCACGACAA ((...(((((((...))))))).((((.(((((..((.....)).))))).....(((((((.((.((((.(.......).)))).)).))))))).))))..))....... ( -28.40) >DroSec_CAF1 5369 112 + 1 GCAAACGGGUUUACAGAACUCGCGCAUUCACACCCGAAUAUCUCUGUGUGUACACUGUACAUCUGCGCGUACAUUACCCGCAUGCACACAUGUACAUAUGCAUGCACGAAUA (((..(((((..........((((((...((((..((.....)).))))((((...))))...))))))......)))))..)))...((((((....))))))........ ( -29.49) >DroEre_CAF1 6459 99 + 1 GCAAACGGGUUUACAGAACUCGCGCAUUCACACCCGAAUAUCUCUGUGUGUACACUGUACAUCUGCGCGUACAU-ACCCGCAUACACACAUGUAUAAAUG------------ .....(((((.(((.(....)(((((...((((..((.....)).))))((((...))))...))))))))...-))))).(((((....))))).....------------ ( -22.40) >consensus GCAAACGGGUUUACAGAACUCGCGCAUUCACACCCGAAUAUCUCUGUGUGUACACUGUACAUCUGCGCGUACAUUACCCGCAUGCACACAUGUACAUAUGCA_GCACGA__A (((..(((((..........((((((...((((..((.....)).))))((((...))))...))))))......)))))..)))........................... (-23.12 = -23.46 + 0.33)


| Location | 16,153,245 – 16,153,357 |
|---|---|
| Length | 112 |
| Sequences | 3 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 89.58 |
| Mean single sequence MFE | -38.53 |
| Consensus MFE | -33.02 |
| Energy contribution | -33.47 |
| Covariance contribution | 0.45 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.77 |
| Structure conservation index | 0.86 |
| SVM decision value | 1.24 |
| SVM RNA-class probability | 0.935309 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16153245 112 - 22224390 UUGUCGUGCUUGCAUAUGUACAUGUGUGCAUGCGGGUAAUGUACGCGCAGAUGUACAGUGUACACACAGAGAUAUUCGGGUGUGAAUGCGCGAGUUCUGUAAACCCGUUUGC ...((((((.(((((.(((((((.(((((.((((.....)))).))))).))))))))))))(((((.(((...)))..)))))...)))))).....(((((....))))) ( -40.90) >DroSec_CAF1 5369 112 - 1 UAUUCGUGCAUGCAUAUGUACAUGUGUGCAUGCGGGUAAUGUACGCGCAGAUGUACAGUGUACACACAGAGAUAUUCGGGUGUGAAUGCGCGAGUUCUGUAAACCCGUUUGC .((((((((((((((.(((((((.(((((.((((.....)))).))))).))))))))))).(((((.(((...)))..))))).))))))))))...(((((....))))) ( -43.90) >DroEre_CAF1 6459 99 - 1 ------------CAUUUAUACAUGUGUGUAUGCGGGU-AUGUACGCGCAGAUGUACAGUGUACACACAGAGAUAUUCGGGUGUGAAUGCGCGAGUUCUGUAAACCCGUUUGC ------------....(((((....))))).((((((-.(((((((...........))))))).(((((...(((((.((......)).))))))))))..)))))).... ( -30.80) >consensus U__UCGUGC_UGCAUAUGUACAUGUGUGCAUGCGGGUAAUGUACGCGCAGAUGUACAGUGUACACACAGAGAUAUUCGGGUGUGAAUGCGCGAGUUCUGUAAACCCGUUUGC ...........((((((....))))))(((.((((((..(((((((...........))))))).(((((...(((((.((......)).))))))))))..)))))).))) (-33.02 = -33.47 + 0.45)

| Location | 16,153,357 – 16,153,456 |
|---|---|
| Length | 99 |
| Sequences | 3 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 79.29 |
| Mean single sequence MFE | -30.35 |
| Consensus MFE | -18.77 |
| Energy contribution | -20.43 |
| Covariance contribution | 1.67 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.39 |
| Structure conservation index | 0.62 |
| SVM decision value | 1.28 |
| SVM RNA-class probability | 0.940185 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16153357 99 - 22224390 CGGCGUGGUCUGCGUA--UGUGUGG-CUCAUACCCAUACACACAUGCAGAUCAAUACACGAACAUAACCUCUAAGCGGUCACUCGCGUAU---GUCUGUAUGUUC .(...((((((((((.--(((((((-.......)))))))...))))))))))...)..(((((((........((((....))))....---.....))))))) ( -32.33) >DroSec_CAF1 5481 102 - 1 CGGCGUGGUCUGCGUA--UGUGUGG-CCCAUACCCAUACACACAUGCAGAUCAAUACACGAACAUAACCUCCAAGCGGUCACUCGCGUAUCGCGUCUGUAUGUUC .(...((((((((((.--(((((((-.......)))))))...))))))))))...)..(((((((........(((((.((....))))))).....))))))) ( -33.82) >DroEre_CAF1 6558 85 - 1 CGGCGUGCCGUGCCUGUGUGUGUGGCCCCAUACCCAUACACACAUUCAGAUCAAUACACGAACAUAACCUCCAUGCGGUCACUCG-------------------- (((.(((((((...(((((((((((........)))))))))))....(.((.......)).)...........)))).))))))-------------------- ( -24.90) >consensus CGGCGUGGUCUGCGUA__UGUGUGG_CCCAUACCCAUACACACAUGCAGAUCAAUACACGAACAUAACCUCCAAGCGGUCACUCGCGUAU___GUCUGUAUGUUC .(...((((((((((...(((((((........)))))))...))))))))))...).(((.....(((.......)))...))).................... (-18.77 = -20.43 + 1.67)


| Location | 16,153,388 – 16,153,496 |
|---|---|
| Length | 108 |
| Sequences | 3 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 88.48 |
| Mean single sequence MFE | -33.20 |
| Consensus MFE | -24.01 |
| Energy contribution | -26.57 |
| Covariance contribution | 2.56 |
| Combinations/Pair | 1.03 |
| Mean z-score | -3.43 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.98 |
| SVM RNA-class probability | 0.893537 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16153388 108 - 22224390 UUUAUAUGCAUCUGUGCAUGUGUGCUAUAUAUGUACAUUCCGGCGUGGUCUGCGUA--UGUGUGG-CUCAUACCCAUACACACAUGCAGAUCAAUACACGAACAUAACCUC ....((((....(((((((((((...)))))))))))...((...((((((((((.--(((((((-.......)))))))...)))))))))).....))..))))..... ( -33.80) >DroSec_CAF1 5515 108 - 1 UUUAUAUGCAUCUGUUCAUGUGUGCUAUAUAUGUACAUUCCGGCGUGGUCUGCGUA--UGUGUGG-CCCAUACCCAUACACACAUGCAGAUCAAUACACGAACAUAACCUC ............(((((..((((((.......))))))...(...((((((((((.--(((((((-.......)))))))...))))))))))...)..)))))....... ( -32.40) >DroEre_CAF1 6572 109 - 1 UUUCAAUGCAUAUGCGCUUGUGUGCUAU--AAGUACAUUCCGGCGUGCCGUGCCUGUGUGUGUGGCCCCAUACCCAUACACACAUUCAGAUCAAUACACGAACAUAACCUC .(((...((((.((((((.((((((...--..))))))...))))))..)))).(((((((((((........)))))))))))...............)))......... ( -33.40) >consensus UUUAUAUGCAUCUGUGCAUGUGUGCUAUAUAUGUACAUUCCGGCGUGGUCUGCGUA__UGUGUGG_CCCAUACCCAUACACACAUGCAGAUCAAUACACGAACAUAACCUC .....((((...(((((((((((...))))))))))).....))))(((((((((...(((((((........)))))))...)))))))))................... (-24.01 = -26.57 + 2.56)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:13:53 2006