
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 16,147,280 – 16,147,374 |
| Length | 94 |
| Max. P | 0.903692 |

| Location | 16,147,280 – 16,147,374 |
|---|---|
| Length | 94 |
| Sequences | 3 |
| Columns | 94 |
| Reading direction | forward |
| Mean pairwise identity | 87.90 |
| Mean single sequence MFE | -19.83 |
| Consensus MFE | -15.79 |
| Energy contribution | -15.80 |
| Covariance contribution | 0.01 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.81 |
| Structure conservation index | 0.80 |
| SVM decision value | 1.03 |
| SVM RNA-class probability | 0.903692 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
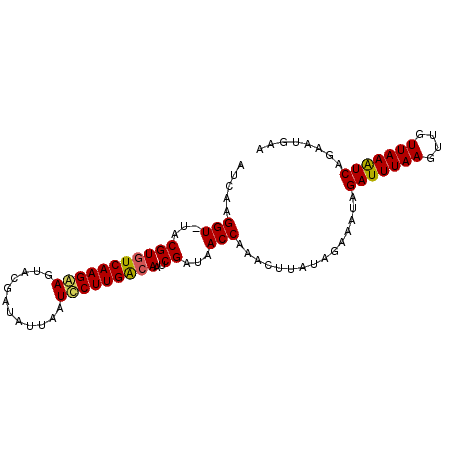
>X_DroMel_CAF1 16147280 94 + 22224390 GUACAGGUAUUCGUGUCAAGGAGUACGAUUUUAAUCCUUGGAAAUCGAUAACCAAACUUAUAGAAAUUGAUUUAAUUUGUUAAGUCAGAAUGAA .....(((..(((..(((((((............)))))))....)))..))).............(((((((((....)))))))))...... ( -20.10) >DroSec_CAF1 3147 93 + 1 AUCAAGGU-UACGUGUCAAGAAGUACGAUAUUAAUCCUUGACAAUCGAUAACCAAAAUUAUAAAAAUAGAUUUAAGUUGUUAAAUCAGAAUGAA .(((.(((-(((((((((((...((......))...)))))))..)).)))))...............(((((((....)))))))....))). ( -18.50) >DroSim_CAF1 2611 93 + 1 AUCAAGGU-UACGUGUCAAGAAGUACAAUAUUAAUUCUUGACAAUCGAUAACCAAACUUAUAGAAAUAGAUUUAAGUUGUUAAAUCAGAAUAAA .((..(((-(((((((((((((............)))))))))..)).)))))...............(((((((....))))))).))..... ( -20.90) >consensus AUCAAGGU_UACGUGUCAAGAAGUACGAUAUUAAUCCUUGACAAUCGAUAACCAAACUUAUAGAAAUAGAUUUAAGUUGUUAAAUCAGAAUGAA .....(((...(((((((((((............)))))))))..))...)))...............(((((((....)))))))........ (-15.79 = -15.80 + 0.01)


| Location | 16,147,280 – 16,147,374 |
|---|---|
| Length | 94 |
| Sequences | 3 |
| Columns | 94 |
| Reading direction | reverse |
| Mean pairwise identity | 87.90 |
| Mean single sequence MFE | -17.27 |
| Consensus MFE | -14.48 |
| Energy contribution | -15.03 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.43 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.90 |
| SVM RNA-class probability | 0.876639 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
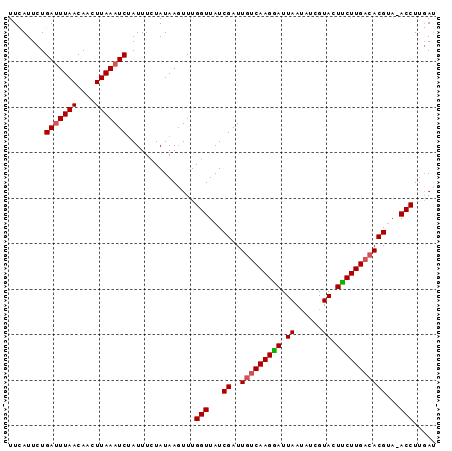
>X_DroMel_CAF1 16147280 94 - 22224390 UUCAUUCUGACUUAACAAAUUAAAUCAAUUUCUAUAAGUUUGGUUAUCGAUUUCCAAGGAUUAAAAUCGUACUCCUUGACACGAAUACCUGUAC ........((((((..(((((.....)))))...)))))).(((..(((.....((((((.((......)).))))))...)))..)))..... ( -13.00) >DroSec_CAF1 3147 93 - 1 UUCAUUCUGAUUUAACAACUUAAAUCUAUUUUUAUAAUUUUGGUUAUCGAUUGUCAAGGAUUAAUAUCGUACUUCUUGACACGUA-ACCUUGAU ........(((((((....)))))))..........(((..(((((.((..(((((((((.((......)).)))))))))))))-)))..))) ( -19.50) >DroSim_CAF1 2611 93 - 1 UUUAUUCUGAUUUAACAACUUAAAUCUAUUUCUAUAAGUUUGGUUAUCGAUUGUCAAGAAUUAAUAUUGUACUUCUUGACACGUA-ACCUUGAU ........(((((((....)))))))...............(((((.((..(((((((((.((......)).)))))))))))))-)))..... ( -19.30) >consensus UUCAUUCUGAUUUAACAACUUAAAUCUAUUUCUAUAAGUUUGGUUAUCGAUUGUCAAGGAUUAAUAUCGUACUUCUUGACACGUA_ACCUUGAU ........(((((((....)))))))...............(((...((..(((((((((.((......)).)))))))))))...)))..... (-14.48 = -15.03 + 0.56)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:13:49 2006