| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 16,140,946 – 16,141,052 |
| Length | 106 |
| Max. P | 0.811663 |

| Location | 16,140,946 – 16,141,052 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 73.10 |
| Mean single sequence MFE | -37.75 |
| Consensus MFE | -15.40 |
| Energy contribution | -15.57 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.35 |
| Mean z-score | -2.05 |
| Structure conservation index | 0.41 |
| SVM decision value | 0.65 |
| SVM RNA-class probability | 0.811663 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16140946 106 + 22224390 AAU---GAGU------UUACUGGU---UGAGCUUGUUCUUGAGAAUGCUCAACUGGGUGAGGCAGUUGUCGCGGUAGUUGCGGCGUUGGGCCAACUGAUCCAGCGGCACCAUCUCGCC ...---....------...(..((---(((((..((((....)))))))))))..)((((((..((.(((((((((((((.(.(....).)))))))..)).)))))))..)))))). ( -40.40) >DroVir_CAF1 3 102 + 1 -----UUAU-------U----UGUUAUUGAGCUUGUUCUUCAAUAAGCUCAGAUGAGUCAGACAGUUGUCACGGUACGUUCGGCGUUCCACCAGCUUGUCCAGCGGCACCAUUUCAUU -----....-------.----......((((((((((....))))))))))((((((...(((....)))..(((.((((.((((..(.....)..)))).))))..)))..)))))) ( -27.50) >DroPse_CAF1 2646 110 + 1 AGUGG-CAU-------UUACUGAUUAUUCAGCUUACUCUUCAGUAUGCUCAAAUGGGUCAGGCAAUUGUCGCGAUAGGUGCGUCGCUCGACCAGCUUGUCGAGCGGCACCAUCUCGUU .((((-((.-------((.((((((.((.(((.((((....)))).))).))...))))))..)).))))))(((.(((((..((((((((......))))))))))))))))..... ( -43.60) >DroSec_CAF1 3592 105 + 1 AAU---GAU-------UUACUGGU---UGAGCUUGCUCUUGAGAAUGCUCAACUGGGUGAGGCAGUUGUCGCGGUAGUUGCGGCGUUGGGCCAACUGAUCCAGCGGCACCAUCUCGCC ...---...-------...(..((---(((((...((....))...)))))))..)((((((..((.(((((((((((((.(.(....).)))))))..)).)))))))..)))))). ( -40.40) >DroAna_CAF1 21147 112 + 1 AAU---UCGUCUACUGUUUCUUAU---UGAGUUGGCUCUUAAGAAUGCUAAGUUGGGUGAGACAGUUGUCCCGGUAGUUGCGGCGGGCGACCAACUGGUCCAAGGGCACCAUUUCGUC ...---..((((.(((((((...(---..(.(((((..........))))).)..)..)))))))(((..(((((.(((((.....)))))..)))))..)))))))........... ( -35.70) >DroPer_CAF1 1549 110 + 1 AGUGG-CAU-------UUACUGAUUAUUCAGCUUACUCUUCAGUAUGCUCAAAUGGGUCAGGCAAUUGUCGCGAUAAGUGCGUCGCUCGACCAGCUUGUCGAGCGGCACCAUCUCGUU .((((-((.-------((.((((((.((.(((.((((....)))).))).))...))))))..)).))))))(((..((((..((((((((......)))))))))))).)))..... ( -38.90) >consensus AAU___CAU_______UUACUGAU___UGAGCUUGCUCUUCAGAAUGCUCAAAUGGGUCAGGCAGUUGUCGCGGUAGGUGCGGCGUUCGACCAACUGGUCCAGCGGCACCAUCUCGUC .............................(((...((....))...)))....(((....(((....)))...........(.((((((((......)))))))).).)))....... (-15.40 = -15.57 + 0.17)

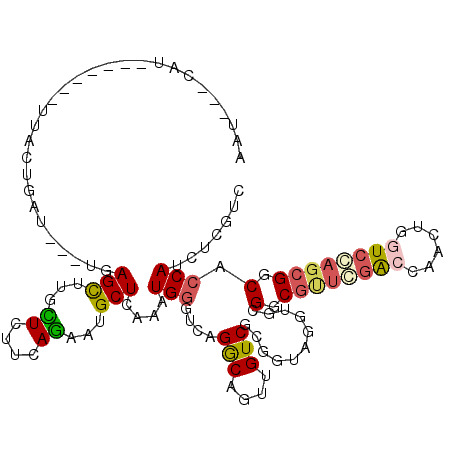

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:13:47 2006