| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 16,132,703 – 16,132,805 |
| Length | 102 |
| Max. P | 0.980865 |

| Location | 16,132,703 – 16,132,805 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 72.62 |
| Mean single sequence MFE | -27.60 |
| Consensus MFE | -20.65 |
| Energy contribution | -20.52 |
| Covariance contribution | -0.13 |
| Combinations/Pair | 1.33 |
| Mean z-score | -1.67 |
| Structure conservation index | 0.75 |
| SVM decision value | 1.87 |
| SVM RNA-class probability | 0.980865 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16132703 102 + 22224390 CGGGCUUAUCCGGCAAGAGUUCACCAUCUCUGGACGAUUCCUCGCAUGAGGAACUCACGCUGGAUGAAAUCAAGGUAAGUUUAUU-------ACCAAAAUAGCCGGGUA (.((((((((((((..((((((..(((....(((....)))....)))..))))))..)))))))))......(((((.....))-------)))......))).)... ( -38.30) >DroGri_CAF1 8181 101 + 1 CUAGCUCAUCUGGCAAAAGCUCUCCAUCACUGGAGGAUUCUUCACACGAGGAACUAGCACUAGACGAAAUCAGGGUAAGUAUGUACAUUUCAAACUAAC----AA---- .....((.(((((.......((((((....)))))).((((((....))))))......))))).)).......((.(((.((.......)).))).))----..---- ( -19.20) >DroSec_CAF1 5786 102 + 1 CGGGCUUAUCCGGCAAGAGUUCACCAUCUCUGGACGAUUCCUCGCAUGAGGAACUCACGCUGGAUGAAAUCAAGGUAAGUUUAUU-------AACAAAAUAGCCGGGUA (.((((((((((((..((((((..(((....(((....)))....)))..))))))..)))))))))...........(((....-------)))......))).)... ( -33.40) >DroSim_CAF1 5799 102 + 1 CGGGCUUAUCCGGCAAGAGUUCACCGUCUCUGGACGAUUCUUCGCAUGAGGAACUCACGCUGGAUGAAAUCAAGGUAAGUUUAUU-------AACAAAAUAGCCGGGUA (.((((((((((((..((((((..((((....)))).(((.......)))))))))..)))))))))...........(((....-------)))......))).)... ( -32.60) >DroEre_CAF1 5807 82 + 1 CGGGCUUAUCCGGCAAGAGUUCACCAUCUCUGGACGAUUCCUCGCAUGAGGAACUCACGUUGGAUGAAAUAAAGGUACG---------------------------GUA ((..((((((((((..((((((..(((....(((....)))....)))..))))))..)))))))).......))..))---------------------------... ( -24.91) >DroAna_CAF1 17003 99 + 1 CUUCUUCAUCUGGAAAGAGUUCACCUUCAUUAGAGGAUUCUUCGCACGAGGAACUUACCUUAGACGAAGUUCGGGUAUGUUUUUA-------A--AAAA-UGACAAAUA .......(((((((.........((((.....))))...(((((...((((......))))...)))))))))))).((((....-------.--....-.)))).... ( -17.20) >consensus CGGGCUUAUCCGGCAAGAGUUCACCAUCUCUGGACGAUUCCUCGCAUGAGGAACUCACGCUGGAUGAAAUCAAGGUAAGUUUAUU_______AACAAAA_AGCCGGGUA .....(((((((((..((((((..(((....(((....)))....)))..))))))..))))))))).......................................... (-20.65 = -20.52 + -0.13)

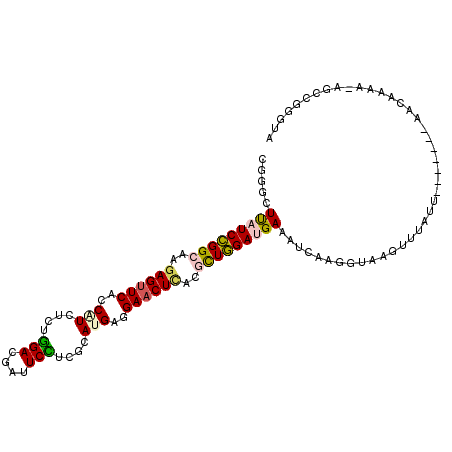
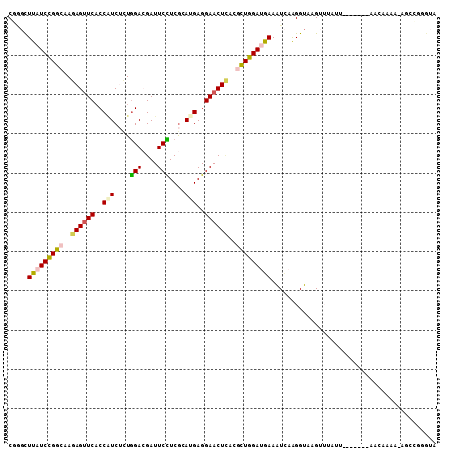
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:13:43 2006