
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 16,132,468 – 16,132,628 |
| Length | 160 |
| Max. P | 0.612982 |

| Location | 16,132,468 – 16,132,588 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 80.39 |
| Mean single sequence MFE | -51.08 |
| Consensus MFE | -31.11 |
| Energy contribution | -32.58 |
| Covariance contribution | 1.48 |
| Combinations/Pair | 1.34 |
| Mean z-score | -1.58 |
| Structure conservation index | 0.61 |
| SVM decision value | 0.02 |
| SVM RNA-class probability | 0.544522 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
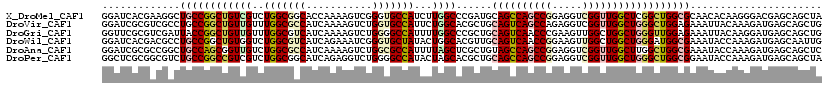
>X_DroMel_CAF1 16132468 120 + 22224390 GGAUCACGAAGGCUGCCGGCUGUCGUCUGGCGGCACCAAAAGUCGGGUGCCAUCUUGGCCCGAUGCAGCCAGCCGGAGGUCGGUUGGCUCGGCUGGCGCAACACAAGGGACGAGCAGCUA ((....(....)...))(((((((((((...((((((........))))))..((((....(.(((.((((((((..((((....))))))))))))))).).)))))))))).))))). ( -55.70) >DroVir_CAF1 7057 120 + 1 GGAUCGCGUCGCCUGCCGGCUGUUGUUUGGCGCCAUCAAAAGUCUGGUGCCAUUCUGGCACGCUGCAGUCAGCCAGAGGUCGGUUGGCUGGGCUGGAGAAAUUACAAAGAUGAGCAGCUG ((....((((((((((((((((.....((((((((.........))))))))(((((((..((....))..)))))))..)))))))).))))..((....)).....)))).....)). ( -44.90) >DroGri_CAF1 7982 120 + 1 GGUUCGCGUCGAUUACCGGCUGUUGUUUGGCGUCAUCAAAAGUCUGGGGCCAUUUUGGCCCGCUGCAGUCAACCCGAAGUUGGCUGGCUGGGUUGGAGAAAUUACAAGGAUGAGCAGCUG ((((((...)))..)))(((((((...((((.(((.........))).))))(((..((((((..((((((((.....)))))))))).))))..)))..............))))))). ( -41.90) >DroWil_CAF1 6353 120 + 1 GGAUCACGACGCCUGCCGGCUGUGGUCUGGCGUCAUCAGAAAUCGGGUGCUAUACUGGCACGUUGCAGUCAACCGGAAGUUGGCUGGCUGGGAUGGCGAAAUACCAAAGAUGAGCAAUUG ((((((((..(((....))))))))))).((.(((((.(....)((.((((((.(..((......(((((((.(....))))))))))..).)))))).....))...)))))))..... ( -42.70) >DroAna_CAF1 16768 120 + 1 GGAUCGCGCCGGCUGCCAGCGGUUGUCUGGCGCCAUCAAAAGUCUGGCGCCAUUUUAGCUCGCUGUAGCCAGCCGGAGGUCGGUUGGCUUGGCUGGCGAAAUACCAAAGAUGAGCAGCUC ((((..((((((((..(((((((((..((((((((.........))))))))...)))).))))).(((((((((.....))))))))).))))))))..)).))............... ( -58.80) >DroPer_CAF1 7641 120 + 1 GGCUCGCGGCGUCUGCCGGCCGUCGUCUGGCGGCAUCAGAGGUCUGGGGCCAUACUAGCACGCUGCAGCCAGCCGGAGGUCGGUUGGCUGGGCUGGCGGAAUACCAAAGAUGAGCAGCUA ((((.((..(((((((((((((.(((((((.(((..(((....)))..)))...)))).))))..((((((((((.....)))))))))))))))))((....))..))))).)))))). ( -62.50) >consensus GGAUCGCGACGCCUGCCGGCUGUUGUCUGGCGCCAUCAAAAGUCUGGUGCCAUUCUGGCACGCUGCAGCCAGCCGGAGGUCGGUUGGCUGGGCUGGCGAAAUACCAAAGAUGAGCAGCUG .............((((((((((((..(((((((...........)))))))...))))......((((((((((.....))))))))))))))))))...................... (-31.11 = -32.58 + 1.48)
| Location | 16,132,508 – 16,132,628 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 80.94 |
| Mean single sequence MFE | -48.55 |
| Consensus MFE | -30.05 |
| Energy contribution | -29.62 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.45 |
| Mean z-score | -1.72 |
| Structure conservation index | 0.62 |
| SVM decision value | 0.16 |
| SVM RNA-class probability | 0.612982 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
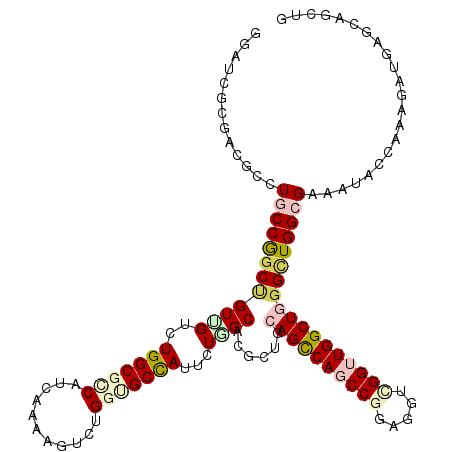
>X_DroMel_CAF1 16132508 120 + 22224390 AGUCGGGUGCCAUCUUGGCCCGAUGCAGCCAGCCGGAGGUCGGUUGGCUCGGCUGGCGCAACACAAGGGACGAGCAGCUACUCAAGGCACUCGCCGAUGCCUGUGCCUUUGACAGGGGCG .(.((((((((.......(((..(((.((((((((..((((....)))))))))))))))......)))..(((......)))..)))))))).)...((((.((.......)).)))). ( -52.60) >DroVir_CAF1 7097 120 + 1 AGUCUGGUGCCAUUCUGGCACGCUGCAGUCAGCCAGAGGUCGGUUGGCUGGGCUGGAGAAAUUACAAAGAUGAGCAGCUGCUGAAAGCGCUAGCAGAUGCCUGUGCCUUUGACAGAGGUG ..(((((((((.....)))))(((.(((((((((.......))))))))))))...........(((((((..(((.((((((.......)))))).)))..))..))))).)))).... ( -44.80) >DroGri_CAF1 8022 120 + 1 AGUCUGGGGCCAUUUUGGCCCGCUGCAGUCAACCCGAAGUUGGCUGGCUGGGUUGGAGAAAUUACAAGGAUGAGCAGCUGCUGAAAGCAUUGGCAGAUGCUUGUGCCUUCGACAGAGGGG .(((..((((..(((..((((((..((((((((.....)))))))))).))))..)))...........(..((((.((((..(.....)..)))).))))..)))))..)))....... ( -49.10) >DroWil_CAF1 6393 120 + 1 AAUCGGGUGCUAUACUGGCACGUUGCAGUCAACCGGAAGUUGGCUGGCUGGGAUGGCGAAAUACCAAAGAUGAGCAAUUGCUCAAAGCUUUGGCCGAUGCGUGUGCAUUUGAUAGAGGAG .(((((((((.((((..((.((((.((((((.(((.....))).)))))).))))))......((((((.(((((....)))))...)))))).......)))))))))))))....... ( -45.20) >DroAna_CAF1 16808 120 + 1 AGUCUGGCGCCAUUUUAGCUCGCUGUAGCCAGCCGGAGGUCGGUUGGCUUGGCUGGCGAAAUACCAAAGAUGAGCAGCUCCUCAAGGCGCUGACCGAUGCCUGUGCCUUUGACAGGGGUG .(((.(((((((((((.(((.((((.(((((((((.....))))))))))))).))))))))........((((......)))).)))))))))..((.(((((.......))))).)). ( -49.80) >DroPer_CAF1 7681 120 + 1 GGUCUGGGGCCAUACUAGCACGCUGCAGCCAGCCGGAGGUCGGUUGGCUGGGCUGGCGGAAUACCAAAGAUGAGCAGCUACUGAAAGCCUUGGCCGAUGCCUGUGCCUUUGACAGAGGCG ((.(..((((.....(((...(((((.(((((((...((((....)))).)))))))((....))........)))))..)))...))))..)))...((((.((.......)).)))). ( -49.80) >consensus AGUCUGGUGCCAUUCUGGCACGCUGCAGCCAGCCGGAGGUCGGUUGGCUGGGCUGGCGAAAUACCAAAGAUGAGCAGCUGCUCAAAGCACUGGCCGAUGCCUGUGCCUUUGACAGAGGCG .(((((..(((.....)))..(((((.(((((((...((((....)))).)))))))(......)........)))))...............)))))((((.((.......)).)))). (-30.05 = -29.62 + -0.44)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:13:42 2006