
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 16,128,168 – 16,128,269 |
| Length | 101 |
| Max. P | 0.961743 |

| Location | 16,128,168 – 16,128,269 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 103 |
| Reading direction | forward |
| Mean pairwise identity | 75.77 |
| Mean single sequence MFE | -29.33 |
| Consensus MFE | -21.50 |
| Energy contribution | -20.53 |
| Covariance contribution | -0.97 |
| Combinations/Pair | 1.35 |
| Mean z-score | -1.72 |
| Structure conservation index | 0.73 |
| SVM decision value | 1.53 |
| SVM RNA-class probability | 0.961743 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16128168 101 + 22224390 GGCAAGUCAUCCUCGGAUUUACCCAGAUGGUGCAGAGGAUGCGCGAAUCGCUUGCCAUCGGCGGUCAAAAAGUUGACGUUCUCUGCAA-AAUUUCG-AAAAUA ((.(((((.......))))).)).((((..(((((((((((.((((.....))))))))....(((((....)))))...))))))).-.))))..-...... ( -29.00) >DroPse_CAF1 1952 96 + 1 GGCAGGUCGUCCUCCGACUUUCCCAAAUGAUGCAGCGGAUGGGCAAAUCUUUUGCCAUCUGCCGUCAGGAAGUUGACAUUCUCUGCAA--A----G-ACCAUA .((((((((.....))))(((((....(((((..((((((((.(((.....)))))))))))))))))))))..........))))..--.----.-...... ( -30.60) >DroWil_CAF1 1063 101 + 1 GGUAAAUCAUCUUCAGAUUUUCCUAAAUGGUGUAGGGGAUGGGCAAAUCGUUUACCAUCUGCAGUAAGGAAAUUAACGUUCUCUGCAA-AUAUAUA-AAGAUC ((.(((((.......))))).)).......(((((((((((......((.(((((........)))))))......))))))))))).-.......-...... ( -20.50) >DroMoj_CAF1 1891 102 + 1 GGUAGAUCAUCUUCGGAUUUGCCCAAGUGGUGUAGCGGAUGCGCAAACCGUUUGCCGUCCGCAGUUAGGAAAUUGACAUUCUCUGCA-UGGAAUAAACAAAUA ((((((((.......))))))))....(.((((((((((((.((((.....))))))))))).(((((....)))))......))))-).)............ ( -31.30) >DroAna_CAF1 5714 97 + 1 GGUAAGUCAUCCUCUGAUUUCCCCAAAUGGUGUAGAGGAUGGGCAAAUCUUUUGCCAUCGGCAGUCAAAAAGUUGACGUUCUCUGCAG-CAUUG-----AAUU ((.((((((.....)))))).))..((((.((((((((((((((((.....))))).(((((.........)))))))))))))))).-)))).-----.... ( -34.00) >DroPer_CAF1 1966 97 + 1 GGCAGGUCGUCCUCCGACUUUCCCAAAUGAUGCAGCGGAUGGGCAAAUCUUUUGCCAUCUGCCGUCAGGAAGUUGACAUUCUCUGCAA-AA----G-ACCAUA .((((((((.....))))(((((....(((((..((((((((.(((.....)))))))))))))))))))))..........))))..-..----.-...... ( -30.60) >consensus GGCAAGUCAUCCUCCGAUUUUCCCAAAUGGUGCAGCGGAUGGGCAAAUCGUUUGCCAUCUGCAGUCAGGAAGUUGACAUUCUCUGCAA_AA_U__G_AAAAUA ((.(((((.......))))).)).......(((((.(((((.((((.....)))))))))...(((((....))))).....)))))................ (-21.50 = -20.53 + -0.97)

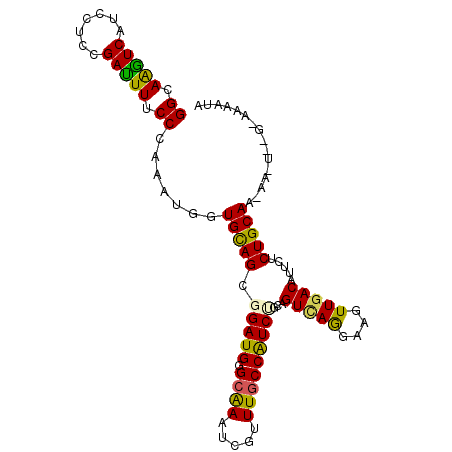

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:13:36 2006