
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 16,122,931 – 16,123,117 |
| Length | 186 |
| Max. P | 0.890918 |

| Location | 16,122,931 – 16,123,021 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 94 |
| Reading direction | reverse |
| Mean pairwise identity | 79.94 |
| Mean single sequence MFE | -25.08 |
| Consensus MFE | -14.12 |
| Energy contribution | -16.07 |
| Covariance contribution | 1.95 |
| Combinations/Pair | 1.23 |
| Mean z-score | -1.88 |
| Structure conservation index | 0.56 |
| SVM decision value | 0.18 |
| SVM RNA-class probability | 0.619323 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16122931 90 - 22224390 GGUGGGGGGACAGCGAAAUUAUUAUAUCCCUGUCCCGAACCACCACCAUAACCGCCACCUAGAGUGCACAUAUUUCAUCGUUUUCAA----UUC (((((.(((((((.((.((....)).)).)))))))...))))).........((.((.....))))....................----... ( -24.20) >DroSim_CAF1 2401 94 - 1 GGUGGGGGGACAGCGAAAUUAUUAUAUCCCUGUCCCGAACCACCACCAUAACCGCCACCUAGAGUGCACAUAUUUCAGCGUUUUCAAUGUAUUC (((((.(((((((.((.((....)).)).)))))))...))))).................(((((((...................))))))) ( -26.01) >DroEre_CAF1 2371 94 - 1 GGUGGGGGGACAGCGAAAUUAUUAUAACCCUGUCCGGCACCACCACCAUAACCGCCACCUAGAGUGCACAUAUUUCAGUGUUUUCAGUGUAUUC (((((((((((((.(.............)))))))((.....)).......)).)))))..((((((((.................)))))))) ( -27.75) >DroWil_CAF1 1843 88 - 1 GGUGGAGGAACAGCAAAGUUGUUGUAUCCCUGUCCGGCACCACCACCGUAACCGCCACCUAAGG---ACAUCAUUAUGUACUU---AUGCAUUU (((((.(((((((((....)))))).))).....(((........)))...)))))...((((.---((((....)))).)))---)....... ( -22.50) >DroYak_CAF1 2323 94 - 1 GGUGGGGGGACAGCGAAAUUGUUAUAUCCCUGUCCGGCACCACCACCAUAACCGCCACCUAGAGUGCACAUAUUUCAGUGUUUUCAGUGUAUUC (((((((((((((.((.((....)).)).))))))((.....)).......)).)))))..((((((((.................)))))))) ( -30.03) >DroPer_CAF1 2077 86 - 1 GGUGGAGGGACAUUAAAAUUAUUAAAGCCCUGGCCG---CCGCCACCAUAACCUCCACCUAAAGCACAAAUAUUUCGAUGUUUUUUU----UU- ((((((((....((((.....)))).....((((..---..))))......)))))))).(((((((.........).))))))...----..- ( -20.00) >consensus GGUGGGGGGACAGCGAAAUUAUUAUAUCCCUGUCCGGCACCACCACCAUAACCGCCACCUAGAGUGCACAUAUUUCAGUGUUUUCAAUGUAUUC (((((..((((((.((.((....)).)).))))))....))))).................(((.((((........)))).)))......... (-14.12 = -16.07 + 1.95)



| Location | 16,122,965 – 16,123,061 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 84.58 |
| Mean single sequence MFE | -30.00 |
| Consensus MFE | -21.26 |
| Energy contribution | -20.71 |
| Covariance contribution | -0.55 |
| Combinations/Pair | 1.29 |
| Mean z-score | -2.52 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.22 |
| SVM RNA-class probability | 0.638767 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16122965 96 + 22224390 GGCGGUUAUGGUGGUGG---------UUCGGGACAGGGAUAUAAUAAUUUCGCUGUCCCCCCACCAAAUUACCAGCAAAUGCCAAAUAAAACGGGUAACUACAAC .(((((.....((((((---------...(((((((.((..........)).))))))).))))))....))).))...((((..........))))........ ( -33.40) >DroVir_CAF1 2 101 + 1 -GCGGUUAUGGUGGCGGUGGCGGUGGUGCCGGACAG---UACAAUAAUUUUGCUGUUCCUCCGCCAAAUUACCAACAAAUGCCAAACAAAACUGGUAAUUAUAAU -..(((.....((((((.(((((.....)))(((((---((.........))))))))).))))))....)))......(((((........)))))........ ( -32.20) >DroEre_CAF1 2409 96 + 1 GGCGGUUAUGGUGGUGG---------UGCCGGACAGGGUUAUAAUAAUUUCGCUGUCCCCCCACCAAACUACCAACAAAUGCCAAAUAAAACGGGUAACUACAAC ((((....(((((((((---------((..((((((.(..((....))..).))))))...))))..))))))).....))))...................... ( -31.00) >DroWil_CAF1 1875 96 + 1 GGCGGUUACGGUGGUGG---------UGCCGGACAGGGAUACAACAACUUUGCUGUUCCUCCACCAAAUUACCAACAAAUGCCAAAUAAAAGUGGUAACUACAAU ...(((.....((((((---------....((((((.((..........)).))))))..))))))....)))......(((((........)))))........ ( -26.00) >DroYak_CAF1 2361 96 + 1 GGCGGUUAUGGUGGUGG---------UGCCGGACAGGGAUAUAACAAUUUCGCUGUCCCCCCACCAAACUACCAACAAAUGCCAAAUAAAACGGGUAACUACAAC ((((....(((((((((---------((..((((((.((..........)).))))))...))))..))))))).....))))...................... ( -32.30) >DroAna_CAF1 1 96 + 1 GGCGGUUAUGGUGGUGG---------UGCCGGGCAAGGCUACAACAAUUUUGCUGUUCCCCCGCCAAACUAUCAACAAAUGCCAAAUAAAUCUGGAAACUAUAAU ((((....(((((((((---------((..(((...(((............)))...))).))))..))))))).....))))..........(....)...... ( -25.10) >consensus GGCGGUUAUGGUGGUGG_________UGCCGGACAGGGAUACAACAAUUUCGCUGUCCCCCCACCAAACUACCAACAAAUGCCAAAUAAAACGGGUAACUACAAC ((..(((.((((((.((.............((((((.(............).)))))))))))))))))..))......((((..........))))........ (-21.26 = -20.71 + -0.55)


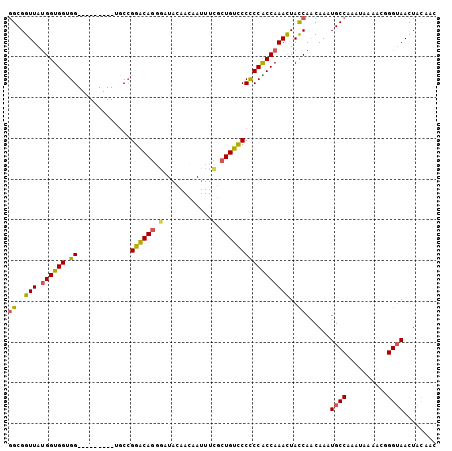
| Location | 16,123,021 – 16,123,117 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 75.11 |
| Mean single sequence MFE | -19.10 |
| Consensus MFE | -12.08 |
| Energy contribution | -12.36 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.63 |
| Structure conservation index | 0.63 |
| SVM decision value | 0.96 |
| SVM RNA-class probability | 0.890918 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
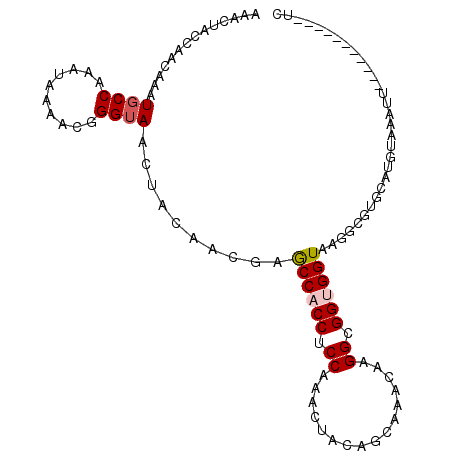
>X_DroMel_CAF1 16123021 96 + 22224390 AAAUUACCAGCAAAUGCCAAAUAAAACGGGUAACUACAACGAGCCACCUCCGAACUACGGCAAACAAGGCGGUGGUAAGGCGUGCAUGCGAACU-----------UU .........(((.(((((........((...........)).((((((.((...(....).......)).))))))..)))))...))).....-----------.. ( -23.50) >DroVir_CAF1 63 106 + 1 AAAUUACCAACAAAUGCCAAACAAAACUGGUAAUUAUAAUGAGCCCCCUCCAAAUUAUAACAAACCAGGCGGUGGUAAGUCCAAUU-GUAAAUCAAACAGAAACUGU ...((((((.....((((.........((((..((((((((((....)))...)))))))...)))))))).))))))........-.........((((...)))) ( -16.90) >DroSim_CAF1 2495 96 + 1 AAACUACCAGCAAAUGCCAAAUAAAACGGGUAACUACAACGAGCCACCUCCGAACUACGGCAAACAAGGCGGUGGUAAGGCGUGCAUGCAAAUU-----------UC .........(((.(((((........((...........)).((((((.((...(....).......)).))))))..)))))...))).....-----------.. ( -23.50) >DroEre_CAF1 2465 96 + 1 AAACUACCAACAAAUGCCAAAUAAAACGGGUAACUACAACGAACCACCUCCAAACUACGGCAAACAAGGCGGUGGUAAGGCGUGCAUGUGAACU-----------CC .........(((.(((((........((...........)).((((((.((................)).))))))..)))))...))).....-----------.. ( -19.79) >DroMoj_CAF1 78 107 + 1 AAAUUACCAGCAAAUGCCAAAUAAAACUGGUAACUAUAACGAGCCCCCUCCAAACUAUAACAAGCCAGGCGGAGGUAAGUACUAUCCAGAAAUUCAAUCCAAAUAGC ...(((((((................))))))).....((..(((.((.((...((......))...)).)).)))..))........................... ( -15.29) >DroAna_CAF1 57 91 + 1 AAACUAUCAACAAAUGCCAAAUAAAUCUGGAAACUAUAAUGAACCACCUCCAAACUACAGCAAGCAAGGUGGUGGUAAG-----CACUUAAAUU-----------UU ..............(((...........(....)........((((((.((...((......))...)).))))))..)-----))........-----------.. ( -15.60) >consensus AAACUACCAACAAAUGCCAAAUAAAACGGGUAACUACAACGAGCCACCUCCAAACUACAGCAAACAAGGCGGUGGUAAGGCGUGCAUGUAAAUU___________UC ..............((((..........))))..........((((((.((................)).))))))............................... (-12.08 = -12.36 + 0.28)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:13:33 2006