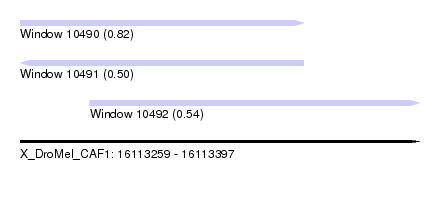
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 16,113,259 – 16,113,397 |
| Length | 138 |
| Max. P | 0.822117 |

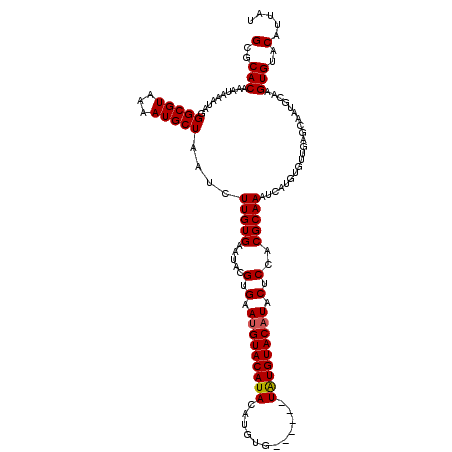
| Location | 16,113,259 – 16,113,357 |
|---|---|
| Length | 98 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 83.62 |
| Mean single sequence MFE | -27.51 |
| Consensus MFE | -17.30 |
| Energy contribution | -18.97 |
| Covariance contribution | 1.67 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.63 |
| Structure conservation index | 0.63 |
| SVM decision value | 0.68 |
| SVM RNA-class probability | 0.822117 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16113259 98 + 22224390 AUAAUGUACACUU----------------GAUUUGCGUGGAGUAUGUACAUA------CACAUGUAUGUACAUUCACGUAUUCACAAGAUUAGCAUUUUACGCCCUAUUUAUUUGUGCGC .....((((((((----------------(...((((((((...((((((((------(....)))))))))))))))))....))))....((.......))..........))))).. ( -28.60) >DroSim_CAF1 1085 114 + 1 AUAAUGUACACUUGCAUUGCUCAACACAUGAUUUGCGUGGAGUAUGUACAUA------CACAUGUAUGUACAUUCCCGUAUUCACAAGAUUAGCAUUUUACGCCCUAUUUAUUUGUGCGC ..((((((....))))))(((.......(((..((((.(((..(((((((((------(....))))))))))))))))).))).......)))......(((.(.........).))). ( -28.24) >DroYak_CAF1 1192 120 + 1 AUAAUGUACACUUGCAUUGCUCAACACAUGAUUUGCGUGGAGUAGGUACACAUGUAUGUACAUAUAUGUACAUUCACAUAUUCACAAGAUUAGCAUUUUACGCCCUAAUUAUUUGUGCGC ..((((((....))))))((.((.((.((((((.((((((((((.((((....)))).)))..((((((......))))))...............)))))))...)))))).)))).)) ( -25.70) >consensus AUAAUGUACACUUGCAUUGCUCAACACAUGAUUUGCGUGGAGUAUGUACAUA______CACAUGUAUGUACAUUCACGUAUUCACAAGAUUAGCAUUUUACGCCCUAUUUAUUUGUGCGC .....(((((...................((..((((((((((((((((((..........)))))))))).)))))))).)).........((.......))..........))))).. (-17.30 = -18.97 + 1.67)



| Location | 16,113,259 – 16,113,357 |
|---|---|
| Length | 98 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 83.62 |
| Mean single sequence MFE | -28.07 |
| Consensus MFE | -16.09 |
| Energy contribution | -16.20 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.51 |
| Structure conservation index | 0.57 |
| SVM decision value | -0.07 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16113259 98 - 22224390 GCGCACAAAUAAAUAGGGCGUAAAAUGCUAAUCUUGUGAAUACGUGAAUGUACAUACAUGUG------UAUGUACAUACUCCACGCAAAUC----------------AAGUGUACAUUAU (..(((..........(((((...))))).......(((...((((.((((((((((....)------)))))))))....))))....))----------------).)))..)..... ( -24.00) >DroSim_CAF1 1085 114 - 1 GCGCACAAAUAAAUAGGGCGUAAAAUGCUAAUCUUGUGAAUACGGGAAUGUACAUACAUGUG------UAUGUACAUACUCCACGCAAAUCAUGUGUUGAGCAAUGCAAGUGUACAUUAU (..(((..........(((((...)))))..((((((....))))))((((((((((....)------))))))))).(((.(((((.....))))).)))........)))..)..... ( -30.40) >DroYak_CAF1 1192 120 - 1 GCGCACAAAUAAUUAGGGCGUAAAAUGCUAAUCUUGUGAAUAUGUGAAUGUACAUAUAUGUACAUACAUGUGUACCUACUCCACGCAAAUCAUGUGUUGAGCAAUGCAAGUGUACAUUAU (..(((.......((((((((...))))...........(((((((.(((((((....))))))).))))))).))))(((.(((((.....))))).)))........)))..)..... ( -29.80) >consensus GCGCACAAAUAAAUAGGGCGUAAAAUGCUAAUCUUGUGAAUACGUGAAUGUACAUACAUGUG______UAUGUACAUACUCCACGCAAAUCAUGUGUUGAGCAAUGCAAGUGUACAUUAU (..(((..........(((((...)))))....(((((.....(.(.(((((((((............))))))))).).)..))))).....................)))..)..... (-16.09 = -16.20 + 0.11)


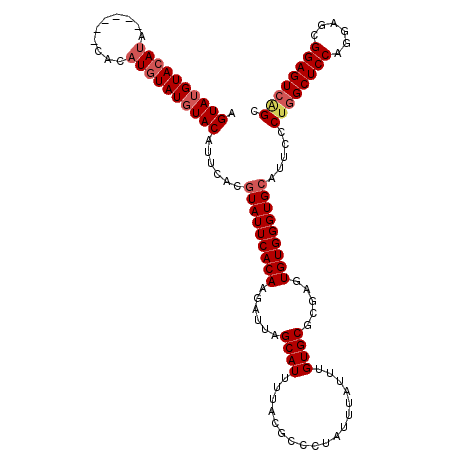
| Location | 16,113,283 – 16,113,397 |
|---|---|
| Length | 114 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 90.40 |
| Mean single sequence MFE | -37.05 |
| Consensus MFE | -27.81 |
| Energy contribution | -29.70 |
| Covariance contribution | 1.89 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.24 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.02 |
| SVM RNA-class probability | 0.541574 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16113283 114 + 22224390 AGUAUGUACAUA------CACAUGUAUGUACAUUCACGUAUUCACAAGAUUAGCAUUUUACGCCCUAUUUAUUUGUGCGCGAGUGUGGGUGCAUUCCCUGGCUCCAGGAGCGGAGUCGGC .(.(((((((((------(....)))))))))).)..(((((((((......((((..................)))).....))))))))).....((((((((......)))))))). ( -38.97) >DroSim_CAF1 1125 114 + 1 AGUAUGUACAUA------CACAUGUAUGUACAUUCCCGUAUUCACAAGAUUAGCAUUUUACGCCCUAUUUAUUUGUGCGCGAGUGUGGGUGCAUUCCCUGGCUCCAGGAGCGGAGUCAGC .(.(((((((((------(....)))))))))).)..(((((((((......((((..................)))).....))))))))).....((((((((......)))))))). ( -38.77) >DroYak_CAF1 1232 120 + 1 AGUAGGUACACAUGUAUGUACAUAUAUGUACAUUCACAUAUUCACAAGAUUAGCAUUUUACGCCCUAAUUAUUUGUGCGCGAGUGUGGGUGCAUUCCCCUGCUCCAGGAGCGGAGUCAGG .(((.((((....)))).)))....((((((...(((((...(((((((((((...........))))))..))))).....))))).))))))...((((((((......)))).)))) ( -33.40) >consensus AGUAUGUACAUA______CACAUGUAUGUACAUUCACGUAUUCACAAGAUUAGCAUUUUACGCCCUAUUUAUUUGUGCGCGAGUGUGGGUGCAUUCCCUGGCUCCAGGAGCGGAGUCAGC .((((((((((..........))))))))))......(((((((((......((((..................)))).....))))))))).....((((((((......)))))))). (-27.81 = -29.70 + 1.89)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:13:27 2006