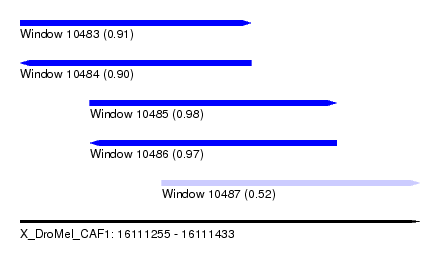
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 16,111,255 – 16,111,433 |
| Length | 178 |
| Max. P | 0.975843 |

| Location | 16,111,255 – 16,111,358 |
|---|---|
| Length | 103 |
| Sequences | 4 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 86.98 |
| Mean single sequence MFE | -35.65 |
| Consensus MFE | -26.17 |
| Energy contribution | -26.93 |
| Covariance contribution | 0.75 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.36 |
| Structure conservation index | 0.73 |
| SVM decision value | 1.10 |
| SVM RNA-class probability | 0.914863 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
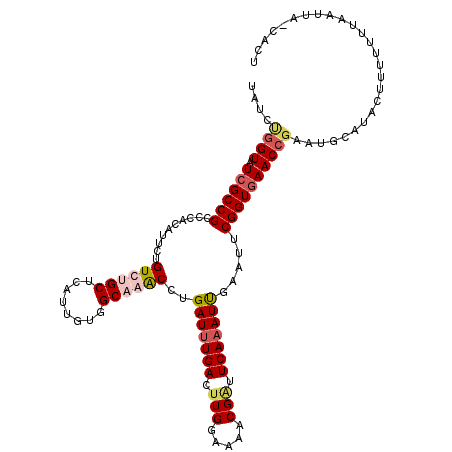
>X_DroMel_CAF1 16111255 103 + 22224390 GCUGGGACACUUCUUCGAUGGAAGGCCGUUGCAUUGAAUCGCUUUCCA--UGCAUUCGGCGACACUAUCUGGUGUCGCCGCCCACAUUCUGUCUGCUCAUUGUAG ..((((...........(((((((((.(((......))).))))))))--).....((((((((((....))))))))))))))........((((.....)))) ( -38.50) >DroSim_CAF1 13755 105 + 1 GCUGGGACACUUCUUCGAUAGAAGGCCUUUGCAUUGAAUCGUUUUCCAAAUGCAUUCGGCGACACUAUCGUGUGUCGCCGCCCACAUUCUGUCUGCUCAUUGUGG ((.(((...(((((.....))))).)))..))...((((((((.....)))).))))((((((((......))))))))..(((((....(....)....))))) ( -35.40) >DroEre_CAF1 25827 105 + 1 GCUGGGAUACUUCUUCGAUGGAAGGCCUUGGUAUUGAAUUGUUUUCCAAAUGUAUCCGGCGACACCAUACGGUAUCGCCGCCCACAUUGUGUCUGCUCAUUGUUG ((..((((((........((((((((..(........)..))))))))(((((...((((((.(((....))).))))))...)))))))))))))......... ( -32.40) >DroYak_CAF1 12279 105 + 1 GCUGGGAAACUUCUUCGAUGGAAGGCCUUCGCAUUGAACUGUUUUCCAAAUGUAUCCGGCGAACCAAUCUGGUAUCGCCGCCCACAUUUUGUCUGCUCCUUGUGG ..((((((((...((((((((((....))).)))))))..))))))))........(((((((((.....))).)))))).(((((....(....)....))))) ( -36.30) >consensus GCUGGGACACUUCUUCGAUGGAAGGCCUUUGCAUUGAAUCGUUUUCCAAAUGCAUCCGGCGACACUAUCUGGUAUCGCCGCCCACAUUCUGUCUGCUCAUUGUGG ..((((...(((((.....)))))......((((((((.....)))..)))))...(((((((((......)))))))))))))........((((.....)))) (-26.17 = -26.93 + 0.75)


| Location | 16,111,255 – 16,111,358 |
|---|---|
| Length | 103 |
| Sequences | 4 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 86.98 |
| Mean single sequence MFE | -33.44 |
| Consensus MFE | -26.16 |
| Energy contribution | -26.85 |
| Covariance contribution | 0.69 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.10 |
| Structure conservation index | 0.78 |
| SVM decision value | 1.01 |
| SVM RNA-class probability | 0.900691 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16111255 103 - 22224390 CUACAAUGAGCAGACAGAAUGUGGGCGGCGACACCAGAUAGUGUCGCCGAAUGCA--UGGAAAGCGAUUCAAUGCAACGGCCUUCCAUCGAAGAAGUGUCCCAGC (((((.((......))...))))).(((((((((......)))))))))..((((--(.(((.....))).)))))..(((((((.......)))).)))..... ( -32.10) >DroSim_CAF1 13755 105 - 1 CCACAAUGAGCAGACAGAAUGUGGGCGGCGACACACGAUAGUGUCGCCGAAUGCAUUUGGAAAACGAUUCAAUGCAAAGGCCUUCUAUCGAAGAAGUGUCCCAGC (((((.((......))...))))).(((((((((......)))))))))..((((((.(((......)))))))))..((((((((.....))))).)))..... ( -36.70) >DroEre_CAF1 25827 105 - 1 CAACAAUGAGCAGACACAAUGUGGGCGGCGAUACCGUAUGGUGUCGCCGGAUACAUUUGGAAAACAAUUCAAUACCAAGGCCUUCCAUCGAAGAAGUAUCCCAGC .........((...(((...))).))(((((((((....)))))))))((((((.(((((..............)))))..((((....))))..)))))).... ( -32.84) >DroYak_CAF1 12279 105 - 1 CCACAAGGAGCAGACAAAAUGUGGGCGGCGAUACCAGAUUGGUUCGCCGGAUACAUUUGGAAAACAGUUCAAUGCGAAGGCCUUCCAUCGAAGAAGUUUCCCAGC ((....)).((.....(((((((..((((((.(((.....)))))))))..)))))))((.((((........((....))((((....))))..)))).)).)) ( -32.10) >consensus CCACAAUGAGCAGACAGAAUGUGGGCGGCGACACCAGAUAGUGUCGCCGAAUACAUUUGGAAAACAAUUCAAUGCAAAGGCCUUCCAUCGAAGAAGUGUCCCAGC .........((......((((((..(((((((((......)))))))))..))))))(((...((........((....))((((....))))..))...))))) (-26.16 = -26.85 + 0.69)



| Location | 16,111,286 – 16,111,396 |
|---|---|
| Length | 110 |
| Sequences | 4 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 85.71 |
| Mean single sequence MFE | -43.92 |
| Consensus MFE | -40.51 |
| Energy contribution | -42.33 |
| Covariance contribution | 1.81 |
| Combinations/Pair | 1.08 |
| Mean z-score | -6.12 |
| Structure conservation index | 0.92 |
| SVM decision value | 1.76 |
| SVM RNA-class probability | 0.975843 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16111286 110 + 22224390 CAUUGAAUCGCUUUCCA--UGCAUUCGGCGACACUAUCUGGUGUCGCCGCCCACAUUCUGUCUGCUCAUUGUAGCCAACCUGAUUUGACUUGGAAAACGAUUCAAAUUGAAU ..((((((((.((((((--..((.(((((((((((....)))))))))..........((.((((.....)))).))....))..))...)))))).))))))))....... ( -40.10) >DroSim_CAF1 13786 112 + 1 CAUUGAAUCGUUUUCCAAAUGCAUUCGGCGACACUAUCGUGUGUCGCCGCCCACAUUCUGUCUGCUCAUUGUGGCCAACCUGAUUUGACUUGGAAAACGAUUCAAAUUGAAU ..((((((((((((((((...((.((((((((((......))))))))(.(((((....(....)....))))).).....))..))..))))))))))))))))....... ( -46.30) >DroEre_CAF1 25858 112 + 1 UAUUGAAUUGUUUUCCAAAUGUAUCCGGCGACACCAUACGGUAUCGCCGCCCACAUUGUGUCUGCUCAUUGUUGCAGACCUGAUAUGACUUGGAAAACGCUUCAAAUUUGAU ..(((((.((((((((((..(((((((((((.(((....))).))))))..........((((((........))))))..)))))...)))))))))).)))))....... ( -42.60) >DroYak_CAF1 12310 112 + 1 CAUUGAACUGUUUUCCAAAUGUAUCCGGCGAACCAAUCUGGUAUCGCCGCCCACAUUUUGUCUGCUCCUUGUGGCAGGCCUGAUUUGACUUGGAAAACGGUUCAAAUCCAAU ..((((((((((((((((.......(((((((((.....))).))))))..((.(((..(((((((......)))))))..))).))..))))))))))))))))....... ( -46.70) >consensus CAUUGAAUCGUUUUCCAAAUGCAUCCGGCGACACUAUCUGGUAUCGCCGCCCACAUUCUGUCUGCUCAUUGUGGCAAACCUGAUUUGACUUGGAAAACGAUUCAAAUUGAAU ..((((((((((((((((.......(((((((((......)))))))))..((.(((..((((((........))))))..))).))..))))))))))))))))....... (-40.51 = -42.33 + 1.81)


| Location | 16,111,286 – 16,111,396 |
|---|---|
| Length | 110 |
| Sequences | 4 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 85.71 |
| Mean single sequence MFE | -47.38 |
| Consensus MFE | -43.59 |
| Energy contribution | -45.02 |
| Covariance contribution | 1.44 |
| Combinations/Pair | 1.14 |
| Mean z-score | -6.17 |
| Structure conservation index | 0.92 |
| SVM decision value | 1.72 |
| SVM RNA-class probability | 0.973950 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16111286 110 - 22224390 AUUCAAUUUGAAUCGUUUUCCAAGUCAAAUCAGGUUGGCUACAAUGAGCAGACAGAAUGUGGGCGGCGACACCAGAUAGUGUCGCCGAAUGCA--UGGAAAGCGAUUCAAUG .......((((((((((((((..(((...(((..(((....))))))...)))...(((((..(((((((((......)))))))))..))))--))))))))))))))).. ( -48.00) >DroSim_CAF1 13786 112 - 1 AUUCAAUUUGAAUCGUUUUCCAAGUCAAAUCAGGUUGGCCACAAUGAGCAGACAGAAUGUGGGCGGCGACACACGAUAGUGUCGCCGAAUGCAUUUGGAAAACGAUUCAAUG .......((((((((((((((..(((...(((..(((....))))))...))).(((((((..(((((((((......)))))))))..))))))))))))))))))))).. ( -50.70) >DroEre_CAF1 25858 112 - 1 AUCAAAUUUGAAGCGUUUUCCAAGUCAUAUCAGGUCUGCAACAAUGAGCAGACACAAUGUGGGCGGCGAUACCGUAUGGUGUCGCCGGAUACAUUUGGAAAACAAUUCAAUA .......(((((..((((((((((((((((...((((((........))))))...)))))..((((((((((....)))))))))).....)))))))))))..))))).. ( -48.70) >DroYak_CAF1 12310 112 - 1 AUUGGAUUUGAACCGUUUUCCAAGUCAAAUCAGGCCUGCCACAAGGAGCAGACAAAAUGUGGGCGGCGAUACCAGAUUGGUUCGCCGGAUACAUUUGGAAAACAGUUCAAUG .......((((((.(((((((..(((......)))((((........))))...(((((((..((((((.(((.....)))))))))..)))))))))))))).)))))).. ( -42.10) >consensus AUUCAAUUUGAAUCGUUUUCCAAGUCAAAUCAGGUCGGCCACAAUGAGCAGACAGAAUGUGGGCGGCGACACCAGAUAGUGUCGCCGAAUACAUUUGGAAAACAAUUCAAUG .......(((((((((((((((...........((((((........))))))..((((((..(((((((((......)))))))))..))))))))))))))))))))).. (-43.59 = -45.02 + 1.44)


| Location | 16,111,318 – 16,111,433 |
|---|---|
| Length | 115 |
| Sequences | 4 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 86.67 |
| Mean single sequence MFE | -26.45 |
| Consensus MFE | -20.80 |
| Energy contribution | -22.43 |
| Covariance contribution | 1.63 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.39 |
| Structure conservation index | 0.79 |
| SVM decision value | -0.03 |
| SVM RNA-class probability | 0.516830 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16111318 115 + 22224390 UAUCUGGUGUCGCCGCCCACAUUCUGUCUGCUCAUUGUAGCCAACCUGAUUUGACUUGGAAAACGAUUCAAAUUGAAUUCGGUGAACCGAAUGCAUACUUUUUUUAAUUACCACU ....(((((((.............((.((((.....)))).))....(((((((.(((.....))).)))))))))((((((....))))))................))))).. ( -23.80) >DroSim_CAF1 13820 115 + 1 UAUCGUGUGUCGCCGCCCACAUUCUGUCUGCUCAUUGUGGCCAACCUGAUUUGACUUGGAAAACGAUUCAAAUUGAAUUCGGUGAACCGAAUGCAUACUUUUUUUAAUUGUCACU ....((((((....(.(((((....(....)....))))).).....(((((((.(((.....))).)))))))..((((((....))))))))))))................. ( -27.10) >DroEre_CAF1 25892 115 + 1 CAUACGGUAUCGCCGCCCACAUUGUGUCUGCUCAUUGUUGCAGACCUGAUAUGACUUGGAAAACGCUUCAAAUUUGAUUCGGUGAACCGAAUUCAAACUUUUUUUAAUUAACACU ....((((.((((((..((.((((.((((((........)))))).)))).))..(((((......)))))........)))))))))).......................... ( -27.20) >DroYak_CAF1 12344 110 + 1 AAUCUGGUAUCGCCGCCCACAUUUUGUCUGCUCCUUGUGGCAGGCCUGAUUUGACUUGGAAAACGGUUCAAAUCCAAUUCGGUAAACCGAAUGCAUACUUUUUUUAA-----ACU .....(((((((((..(((((....(....)....)))))..)))..(((((((.(((.....))).)))))))..((((((....))))))).)))))........-----... ( -27.70) >consensus UAUCUGGUAUCGCCGCCCACAUUCUGUCUGCUCAUUGUGGCAAACCUGAUUUGACUUGGAAAACGAUUCAAAUUGAAUUCGGUGAACCGAAUGCAUACUUUUUUUAAUUA_CACU ....((((.((((((..........((((((........))))))..(((((((.(((.....))).))))))).....)))))))))).......................... (-20.80 = -22.43 + 1.63)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:13:22 2006