| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 16,110,874 – 16,110,994 |
| Length | 120 |
| Max. P | 0.776991 |

| Location | 16,110,874 – 16,110,994 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 81.06 |
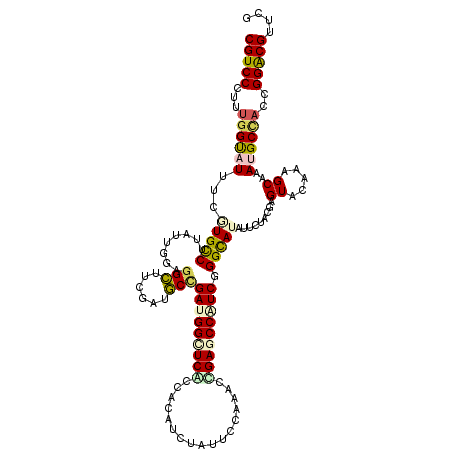
| Mean single sequence MFE | -39.73 |
| Consensus MFE | -28.35 |
| Energy contribution | -28.63 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.40 |
| Mean z-score | -1.74 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.55 |
| SVM RNA-class probability | 0.776991 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
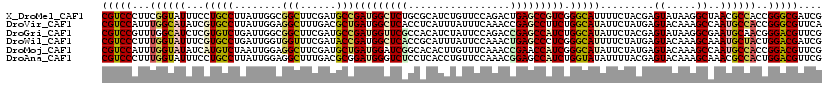
>X_DroMel_CAF1 16110874 120 + 22224390 CGUCCCUUCGGUAUUUCCUGCCUUAUUGGCGGCUUCGAUGCCGAUGGCUCUGCGCAUCUGUUCCAGACUGAGCCGUCGGGCAUUUUCUACGAGUAUAAGGCUAACGCCACCGGGCGAUCG (((((.((((.......(((((.....)))))....((((((((((((((......((((...))))..)))))))).)))))).....)))).....(((....)))...))))).... ( -46.50) >DroVir_CAF1 164520 120 + 1 CGUCCAUUUGGCAUAUCGUGCCUUAUUGGAGGCUUUGACGCUGAUGGCUCACCUCAUUUAUUUCAAACCGAGCCUUCUGGCAUAUUCUAUGAGUACAAAGCCAAUGCCACCGGGCGUUCA ..((((...(((((...)))))....))))(((((((..(((((.(((((...................))))).)).))).(((((...))))))))))))((((((....)))))).. ( -40.21) >DroGri_CAF1 161521 120 + 1 CGUCCGUUUGGCAUCUCGUGUCUGAUUGGCGGCUUCGAUGCCGAUGGUUCGCCACAUCUAUUCCAGACCGAGCCAUCUGGCAUAUUCUACGAGUAUAAGGCGAAUGCAACGGGACGUUCG (((((((...((((.((((.(...(((.(..(.....((((((((((((((.....(((.....))).))))))))).)))))...)..).)))...).)))))))).)).))))).... ( -40.00) >DroWil_CAF1 234181 120 + 1 CGUCCCUUUGGUAUUUCGUGCCUGAUUGGUGGUUUCGAUACCGAUGGCUCACCGCAUUUAUUCCAAACUGAGCCCUCGGGCAUUUUCUAUGAGUACAAAGCAAAUGCUACUGGACGAUCG (((((...((((((((.((((((((((((((.......)))))).((((((.................)))))).)))))))).........((.....))))))))))..))))).... ( -35.93) >DroMoj_CAF1 165249 120 + 1 CGUCCAUUUGGUAUAUCAUGUCUAAUUGGAGGCUUCGAUGCUGAUGGAUCGGCACACUUGUUUCAAACCGAACCAUCGGGCAUAUUCUAUGAGUACAAAGCCAAUGCCACCGGACGUUCG (((((...((((((......(((....)))(((((.(((((((((((.((((...............)))).))))).)))))((((...)))).).))))).))))))..))))).... ( -37.26) >DroAna_CAF1 52700 120 + 1 CGUCCCUUUGGUAUUUCCUGCCUUAUUGGAGGCUUUGACGCGGAUGGGUCUCCUCACCUGUUCCAAACGGAGCCAUCUGGUAUAUUUUACGAGUACAAAGCAAACGCCACUGGACGUUCG (((((...((((..((((.........))))((((((((.(((((((((......))).(((((....)))))))))))(((.....)))..)).))))))....))))..))))).... ( -38.50) >consensus CGUCCCUUUGGUAUUUCGUGCCUUAUUGGAGGCUUCGAUGCCGAUGGCUCACCACAUCUAUUCCAAACCGAGCCAUCGGGCAUAUUCUACGAGUACAAAGCAAAUGCCACCGGACGUUCG (((((...((((((...(((((........(((......)))(((((((((.................))))))))).))))).........((.....))..))))))..))))).... (-28.35 = -28.63 + 0.28)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:13:18 2006