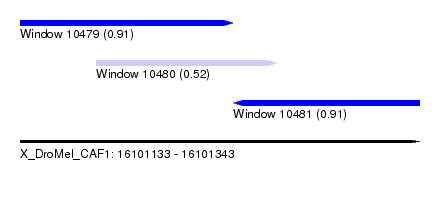
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 16,101,133 – 16,101,343 |
| Length | 210 |
| Max. P | 0.913114 |

| Location | 16,101,133 – 16,101,245 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 90.96 |
| Mean single sequence MFE | -33.56 |
| Consensus MFE | -27.98 |
| Energy contribution | -27.74 |
| Covariance contribution | -0.24 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.72 |
| Structure conservation index | 0.83 |
| SVM decision value | 1.07 |
| SVM RNA-class probability | 0.910801 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16101133 112 + 22224390 UGACUUCAUCGCAACAUGACGAUGAAUAGAUGGCCGAAAGGUUAAUGUUAA--UGUUGGAUAAACAGCGUUCGGAAGAGCUCGUCUUAGAGCUGCUGUUCCGGAAACGAUUGUU ((((((((((((.....).)))))))....(((((....)))))..)))).--....(((...(((((..((....))((((......)))).))))))))(....)....... ( -36.70) >DroSec_CAF1 3041 112 + 1 UGACCUCAUCGCAACAUGAUGAUGAAUAGAAGGCCGAAGGGUUAAUGUUAA--UGUUGGAUAAACAGCGUUCGGAAGAGCUCGUCAUAGAGCUGCUGUUCCGGAAACGAUUGUU .......(((.((((((.......((((...((((....))))..)))).)--)))))))).((((((..((....))((((......)))).)))))).((....))...... ( -32.80) >DroSim_CAF1 1689 112 + 1 UGACCUCAUCGCAACAUGCUGAUGAAUAGAAGGCCGAAGGGUUAAUGUUAA--UGUUGGAUAAACAGCGUUCGGAAGAGCUCGUCUUGGAGCUGCUGUUCCGGAAACGAUUGUU ((((.(((((((.....)).)))))......((((....))))...)))).--.((((......))))((((....))))((((.(((((((....)))))))..))))..... ( -33.90) >DroEre_CAF1 16137 112 + 1 UGACUUCAUCGCAACAUGAUGAUGAAAAGCAGGUCCUAGGGUUAAUGUUAG--AGUUGGAUAAACAGCGUUCGGAAGAGCUCGUGUUGGAGCUGCUGUUCCGGAAACGAUUGUU .....((((((.....)))))).(((.(((((.(((...............--.((((......))))((((....)))).......))).))))).)))((....))...... ( -33.90) >DroYak_CAF1 1703 114 + 1 AGACUUCAUCGCAACAUGAUGAUGAAAAGAAGGCCGUAGGGUUAAUGUUUAUGUGGUGGAUAAACAGCGUUCGGAAUAGCUCGUCUUUGAGCAGCAGUUCCGGAAACGAUUGUU ....(((((((((.(((....))).......((((....))))........)))))))))..(((((((((((((((.(((((....)))))....)))))))..))).))))) ( -30.50) >consensus UGACUUCAUCGCAACAUGAUGAUGAAUAGAAGGCCGAAGGGUUAAUGUUAA__UGUUGGAUAAACAGCGUUCGGAAGAGCUCGUCUUAGAGCUGCUGUUCCGGAAACGAUUGUU ((((.(((((((.....).))))))......((((....))))...)))).......(((...(((((..((....))((((......)))).))))))))(....)....... (-27.98 = -27.74 + -0.24)


| Location | 16,101,173 – 16,101,268 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 99 |
| Reading direction | forward |
| Mean pairwise identity | 84.36 |
| Mean single sequence MFE | -27.05 |
| Consensus MFE | -18.76 |
| Energy contribution | -19.68 |
| Covariance contribution | 0.92 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.09 |
| Structure conservation index | 0.69 |
| SVM decision value | -0.03 |
| SVM RNA-class probability | 0.515961 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16101173 95 + 22224390 GUUAAUGUUAA--UGUUGGAUAAACAGCGUUCGGAAGAGCUCGUCUUAGAGCUGCUGUUCCGGA--AACGAUUGUUUAAAAUUCCUUAGUUUGGCAGCU .....((((((--....(((((((((((((((((((((((((......))))).))..))))).--.))).)))))))....))).....))))))... ( -27.30) >DroSec_CAF1 3081 95 + 1 GUUAAUGUUAA--UGUUGGAUAAACAGCGUUCGGAAGAGCUCGUCAUAGAGCUGCUGUUCCGGA--AACGAUUGUUUAAAAUUCCUUAGUUUGGCAGCU .....((((((--....(((((((((((((((((((((((((......))))).))..))))).--.))).)))))))....))).....))))))... ( -27.30) >DroSim_CAF1 1729 95 + 1 GUUAAUGUUAA--UGUUGGAUAAACAGCGUUCGGAAGAGCUCGUCUUGGAGCUGCUGUUCCGGA--AACGAUUGUUUAAAAUUCCUUAGUUUGGCAGCU .....((((((--....((((((((((.((((....))))((((.(((((((....))))))).--.)))))))))))....))).....))))))... ( -27.30) >DroEre_CAF1 16177 95 + 1 GUUAAUGUUAG--AGUUGGAUAAACAGCGUUCGGAAGAGCUCGUGUUGGAGCUGCUGUUCCGGA--AACGAUUGUUUAAAAUUCCUUAGUUUGGCAGCU .....((((((--(.(.((((((((((.((((....))))((((..((((((....))))))..--.)))))))))))....)))..).)))))))... ( -29.60) >DroYak_CAF1 1743 97 + 1 GUUAAUGUUUAUGUGGUGGAUAAACAGCGUUCGGAAUAGCUCGUCUUUGAGCAGCAGUUCCGGA--AACGAUUGUUUCAAAUUCCUUAGUUUGGCAGCU ((((((((((((.......)))))))......(((((.(((((....)))))((((((..((..--..))))))))....))))).....))))).... ( -24.00) >DroAna_CAF1 33583 90 + 1 ---AUUUUUAG--UGCU-GACCAACAGGGUUCGGAAGAGUUCGCUUCCGAACAACUAUUCUAAAUCAUUAAUUGUUUUAAAUUCUU---UUUGGCAGCU ---........--.(((-(.((((.((((((((((((......)))))))))........((((.((.....)).))))....)))---.)))))))). ( -26.80) >consensus GUUAAUGUUAA__UGUUGGAUAAACAGCGUUCGGAAGAGCUCGUCUUAGAGCUGCUGUUCCGGA__AACGAUUGUUUAAAAUUCCUUAGUUUGGCAGCU .....((((((......(((((((((((((((((((.(((((......)))))....))))))....))).)))))))....))).....))))))... (-18.76 = -19.68 + 0.92)



| Location | 16,101,245 – 16,101,343 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 98 |
| Reading direction | reverse |
| Mean pairwise identity | 91.02 |
| Mean single sequence MFE | -23.93 |
| Consensus MFE | -22.11 |
| Energy contribution | -22.20 |
| Covariance contribution | 0.09 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.48 |
| Structure conservation index | 0.92 |
| SVM decision value | 1.09 |
| SVM RNA-class probability | 0.913114 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16101245 98 - 22224390 AAAGAAAACCACCAGCUUCCCAAAGCCUGAUUACGAAAUCCAGUGUCCUGGGGAGUGGCCUCAUUUGUCGCUCUAAGCUGCCAAACUAAGGAAUUUUA ............((((((((((..((((((((....))).))).))..))))(((((((.......))))))).))))))((.......))....... ( -24.80) >DroSec_CAF1 3153 98 - 1 AAAGAAAACCACCAGCUUCCCAAAACCUGAUUACGAAAUCCAGUGUCGUGGGGAGUGGCCUCAUUUGUCGCUCUAAGCUGCCAAACUAAGGAAUUUUA ............((((((((((..((((((((....))).))).))..))))(((((((.......))))))).))))))((.......))....... ( -25.60) >DroSim_CAF1 1801 98 - 1 AAAGAAAACCACCAGCUUCCCAAAACCUGAUUACGAAAUCCAGUGUCGUGGGGAGUGGCCUCAUUUGUCGCUCUAAGCUGCCAAACUAAGGAAUUUUA ............((((((((((..((((((((....))).))).))..))))(((((((.......))))))).))))))((.......))....... ( -25.60) >DroEre_CAF1 16249 98 - 1 AAAGAAAACCACCAGCUUCCCAAAACCUGAUUACCAAAUCCAGUGUCGUGGGGAGUGGCCUCAUUUGUCGCUCUAAGCUGCCAAACUAAGGAAUUUUA ............((((((((((..((((((((....))).))).))..))))(((((((.......))))))).))))))((.......))....... ( -25.60) >DroYak_CAF1 1817 98 - 1 GAAGAAAACCACCAGCUUCCCAAAACCUGAUUACGAAAUCCAGUGUCGUGGGGAGUGGCCUCAUUUGUCGCUCUAAGCUGCCAAACUAAGGAAUUUGA ............((((((((((..((((((((....))).))).))..))))(((((((.......))))))).))))))((.......))....... ( -25.60) >DroAna_CAF1 33653 92 - 1 AAAGAAAACCACCAGCUUCCUGUAAACUGCGUAGAAA---CAGAGUUAAAGGGAGUUGCCUCAUUUGUCGCUCUAAGCUGCCAAA---AAGAAUUUAA ............(((((((((...((((..((....)---)..))))..))))))))).....((((.(((.....)).).))))---.......... ( -16.40) >consensus AAAGAAAACCACCAGCUUCCCAAAACCUGAUUACGAAAUCCAGUGUCGUGGGGAGUGGCCUCAUUUGUCGCUCUAAGCUGCCAAACUAAGGAAUUUUA ............((((((((((..(((((...........))).))..))))(((((((.......))))))).)))))).................. (-22.11 = -22.20 + 0.09)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:13:17 2006