
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 16,065,745 – 16,065,859 |
| Length | 114 |
| Max. P | 0.999954 |

| Location | 16,065,745 – 16,065,859 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.19 |
| Mean single sequence MFE | -36.24 |
| Consensus MFE | -33.76 |
| Energy contribution | -34.08 |
| Covariance contribution | 0.32 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.10 |
| Structure conservation index | 0.93 |
| SVM decision value | 3.37 |
| SVM RNA-class probability | 0.999089 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16065745 114 + 22224390 ACUCGACUAUCUGGGGAUUAUUUGUGUGGUUUACAGUUUUUAAUUAUGCACCGCGACAUGCGGAUGCAGUGGUAUUAUAAUGGCCACGGAUCCCCAGACCGCAAGUUAAACGAA------ ..(((....((((((((((..(((((.....)))))..........((((((((.....)))).))))(((((.........))))).)))))))))).(....).....))).------ ( -39.00) >DroSec_CAF1 11531 114 + 1 ACUCGACUAUCUGGGGAUUAUUUGUGUGGUUUACAGUUUUUAAUUAUGCACCGCGACAUGCGGAUGCAGUGGUAUUAUAAUGGCCAGGGAUCCCCAGACCGCAAGUUUAACGAG------ .((((....((((((((((..(((((.....)))))..........((((((((.....)))).)))).((((.........))))..)))))))))).(....).....))))------ ( -37.60) >DroSim_CAF1 9423 114 + 1 ACUCGACUAUCUGGGGAUUAUUUGUGUGGUUUACAGUUUUUAAUUAUGCACCGCGACAUGCGGAUGCAGUGGUAUUAUAAUGGCCAGGGAUCCCCAGACCGCAAGUUCAACGAA------ ..(((....((((((((((..(((((.....)))))..........((((((((.....)))).)))).((((.........))))..)))))))))).(....).....))).------ ( -35.40) >DroEre_CAF1 11753 120 + 1 ACUCUACUAUCUGGGGAUUAUUUGUGUGGUUUACAGUUUUUAAUUAUGCACCGCGACAUGCGGAUGCAGUGGUAUUAUAAUGGCCAUGGAUCCCCAGACCGCAAGUUCGACGAAAUUUCA .........((((((((((..(((((.....)))))..........((((((((.....)))).))))(((((.........))))).)))))))))).(....)............... ( -34.60) >DroYak_CAF1 7873 114 + 1 ACUCUACUAUCUGGGGAUUAUUUGUGUGGUUUACAGUUUUUAAUUAUGCACCGCGACAUGCGGAUGCAGUGGUAUUAUAAUGGCCAUGGAUCCCCAGACCGCAAGUUUGACGAA------ .........((((((((((..(((((.....)))))..........((((((((.....)))).))))(((((.........))))).)))))))))).(....).........------ ( -34.60) >consensus ACUCGACUAUCUGGGGAUUAUUUGUGUGGUUUACAGUUUUUAAUUAUGCACCGCGACAUGCGGAUGCAGUGGUAUUAUAAUGGCCAGGGAUCCCCAGACCGCAAGUUCAACGAA______ .........((((((((((..(((((.....)))))..........((((((((.....)))).))))(((((.........))))).)))))))))).(....)............... (-33.76 = -34.08 + 0.32)

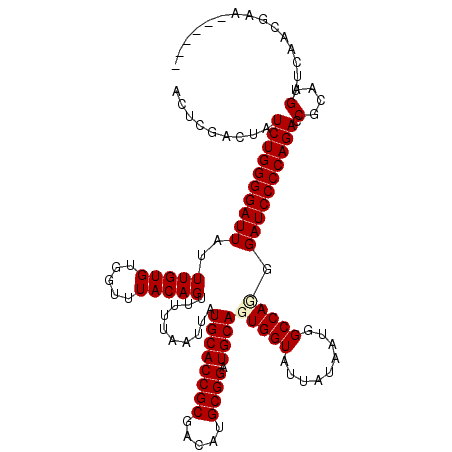

| Location | 16,065,745 – 16,065,859 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.19 |
| Mean single sequence MFE | -33.32 |
| Consensus MFE | -31.58 |
| Energy contribution | -31.98 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.73 |
| Structure conservation index | 0.95 |
| SVM decision value | 4.82 |
| SVM RNA-class probability | 0.999954 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16065745 114 - 22224390 ------UUCGUUUAACUUGCGGUCUGGGGAUCCGUGGCCAUUAUAAUACCACUGCAUCCGCAUGUCGCGGUGCAUAAUUAAAAACUGUAAACCACACAAAUAAUCCCCAGAUAGUCGAGU ------........((((((.((((((((((..((((...........))))((((.((((.....))))))))............................)))))))))).).))))) ( -36.80) >DroSec_CAF1 11531 114 - 1 ------CUCGUUAAACUUGCGGUCUGGGGAUCCCUGGCCAUUAUAAUACCACUGCAUCCGCAUGUCGCGGUGCAUAAUUAAAAACUGUAAACCACACAAAUAAUCCCCAGAUAGUCGAGU ------........((((((.((((((((((...(((...........))).((((.((((.....))))))))............................)))))))))).).))))) ( -33.20) >DroSim_CAF1 9423 114 - 1 ------UUCGUUGAACUUGCGGUCUGGGGAUCCCUGGCCAUUAUAAUACCACUGCAUCCGCAUGUCGCGGUGCAUAAUUAAAAACUGUAAACCACACAAAUAAUCCCCAGAUAGUCGAGU ------........((((((.((((((((((...(((...........))).((((.((((.....))))))))............................)))))))))).).))))) ( -33.20) >DroEre_CAF1 11753 120 - 1 UGAAAUUUCGUCGAACUUGCGGUCUGGGGAUCCAUGGCCAUUAUAAUACCACUGCAUCCGCAUGUCGCGGUGCAUAAUUAAAAACUGUAAACCACACAAAUAAUCCCCAGAUAGUAGAGU ..............((((((.((((((((((...(((...........))).((((.((((.....))))))))............................)))))))))).))).))) ( -31.70) >DroYak_CAF1 7873 114 - 1 ------UUCGUCAAACUUGCGGUCUGGGGAUCCAUGGCCAUUAUAAUACCACUGCAUCCGCAUGUCGCGGUGCAUAAUUAAAAACUGUAAACCACACAAAUAAUCCCCAGAUAGUAGAGU ------........((((((.((((((((((...(((...........))).((((.((((.....))))))))............................)))))))))).))).))) ( -31.70) >consensus ______UUCGUUAAACUUGCGGUCUGGGGAUCCAUGGCCAUUAUAAUACCACUGCAUCCGCAUGUCGCGGUGCAUAAUUAAAAACUGUAAACCACACAAAUAAUCCCCAGAUAGUCGAGU ..............(((((..((((((((((...(((...........))).((((.((((.....))))))))............................))))))))))...))))) (-31.58 = -31.98 + 0.40)

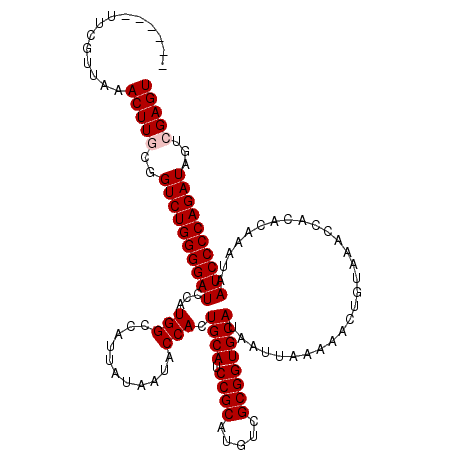

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:13:00 2006