| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 16,051,712 – 16,051,821 |
| Length | 109 |
| Max. P | 0.892796 |

| Location | 16,051,712 – 16,051,821 |
|---|---|
| Length | 109 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.86 |
| Mean single sequence MFE | -32.96 |
| Consensus MFE | -25.84 |
| Energy contribution | -26.24 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.64 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.38 |
| SVM RNA-class probability | 0.711083 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16051712 109 + 22224390 AUGGGGGCGAUUCGCUUCGAUUUU-UUUUCCCUGUCGCCACACAGCACCAAUGGAAUUCGAGCACUGGUGACAAAGAAGCGACGCUGCUGCUUCCUCUUUCCAC--CCCAC--------A .(((((..((.(((((((((....-...))..((((((((....((...............))..))))))))..))))))).((....))........))..)--)))).--------. ( -31.46) >DroSec_CAF1 5907 111 + 1 AUGGGGGCGAUUCGCUUCGAUUUU-UUUUCCCUGUCGCCACACAGCACCAAUGGAAUUCGAGCACUGGUGACAAAGAAGCGACGCUGCUGCUUCCUCUUUCCCCCACCCCC--------A .((((((.(..(((((((((....-...))..((((((((....((...............))..))))))))..))))))).((....)).............).)))))--------) ( -34.06) >DroSim_CAF1 2600 111 + 1 AUGGGGGCGAUUCGCUUCGUUUUU-UUUUCCCUGUCGCCACACAGCACCAAUGGAAUUCGAGCACUGGUGACAAAGAAGCGACGCUGCUGCUUCCUCUUUCCCCCACCCCC--------A .((((((.(..(((((((((((..-..((((.((..((......))..))..))))...))))..((....))..))))))).((....)).............).)))))--------) ( -34.20) >DroEre_CAF1 6398 116 + 1 AUGGGGGCGAUUCGCUUCGAUUUU--UUUCCCUGUCGCCACACAGCACCAAUGGAAUUCGAGCACUGGUGACAAAGAAGCGUCGCUGCUGCUUCCUCUUUCCAC--CCCUCUUUCCACUA ..(((((((((.......((....--..))...))))))............(((((...(((((..((((((........))))))..))))).....))))).--)))........... ( -31.50) >DroYak_CAF1 2336 110 + 1 AUGGGGGCGAUUCGCUUCGAUUUUUUUUUCCCUGUCGCCCCACAGCACCAAUGGAAUUCGAGCACUGGUGACAAAGAAGCGUCGCUGCUGCUUCCUCUUUGCAC--CCCCAU-------- ((((((((((..((((((.............((((......))))(((((.((.........)).))))).....))))))))))...(((.........))).--))))))-------- ( -33.60) >consensus AUGGGGGCGAUUCGCUUCGAUUUU_UUUUCCCUGUCGCCACACAGCACCAAUGGAAUUCGAGCACUGGUGACAAAGAAGCGACGCUGCUGCUUCCUCUUUCCAC__CCCCC________A ..((.((((..(((((((.............((((......))))(((((.((.........)).))))).....))))))))))).))............................... (-25.84 = -26.24 + 0.40)

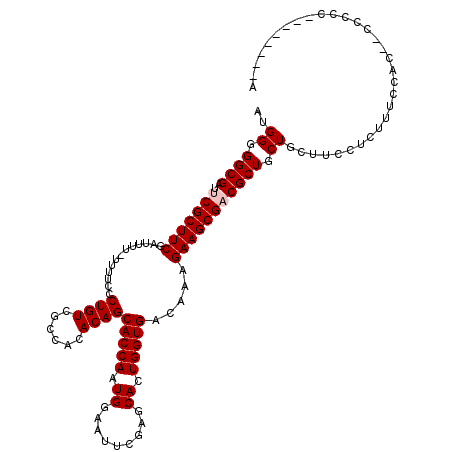

| Location | 16,051,712 – 16,051,821 |
|---|---|
| Length | 109 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.86 |
| Mean single sequence MFE | -41.50 |
| Consensus MFE | -31.00 |
| Energy contribution | -32.00 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.43 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.97 |
| SVM RNA-class probability | 0.892796 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16051712 109 - 22224390 U--------GUGGG--GUGGAAAGAGGAAGCAGCAGCGUCGCUUCUUUGUCACCAGUGCUCGAAUUCCAUUGGUGCUGUGUGGCGACAGGGAAAA-AAAAUCGAAGCGAAUCGCCCCCAU (--------(.(((--((((.........((....)).(((((((.((((((((((..(.(((......))))..))).)))))))....((...-....))))))))).))))))))). ( -40.60) >DroSec_CAF1 5907 111 - 1 U--------GGGGGUGGGGGAAAGAGGAAGCAGCAGCGUCGCUUCUUUGUCACCAGUGCUCGAAUUCCAUUGGUGCUGUGUGGCGACAGGGAAAA-AAAAUCGAAGCGAAUCGCCCCCAU (--------((((((((............((....)).(((((((.((((((((((..(.(((......))))..))).)))))))....((...-....))))))))).))))))))). ( -44.40) >DroSim_CAF1 2600 111 - 1 U--------GGGGGUGGGGGAAAGAGGAAGCAGCAGCGUCGCUUCUUUGUCACCAGUGCUCGAAUUCCAUUGGUGCUGUGUGGCGACAGGGAAAA-AAAAACGAAGCGAAUCGCCCCCAU (--------((((((((............((....)).(((((((.((((((((((..(.(((......))))..))).)))))))..(......-.....)))))))).))))))))). ( -43.60) >DroEre_CAF1 6398 116 - 1 UAGUGGAAAGAGGG--GUGGAAAGAGGAAGCAGCAGCGACGCUUCUUUGUCACCAGUGCUCGAAUUCCAUUGGUGCUGUGUGGCGACAGGGAAA--AAAAUCGAAGCGAAUCGCCCCCAU .........(.(((--((((.........((....))..((((((.((((((((((..(.(((......))))..))).)))))))....((..--....))))))))..)))))))).. ( -39.50) >DroYak_CAF1 2336 110 - 1 --------AUGGGG--GUGCAAAGAGGAAGCAGCAGCGACGCUUCUUUGUCACCAGUGCUCGAAUUCCAUUGGUGCUGUGGGGCGACAGGGAAAAAAAAAUCGAAGCGAAUCGCCCCCAU --------((((((--(((..........((....))..(((((((((((((((((((.........)))))))(((....)))))))))((........))))))))...))))))))) ( -39.40) >consensus U________GGGGG__GUGGAAAGAGGAAGCAGCAGCGUCGCUUCUUUGUCACCAGUGCUCGAAUUCCAUUGGUGCUGUGUGGCGACAGGGAAAA_AAAAUCGAAGCGAAUCGCCCCCAU .........(((((...............((....))..((((((.((((((((((..(.(((......))))..))).)))))))....((........)))))))).....))))).. (-31.00 = -32.00 + 1.00)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:12:56 2006