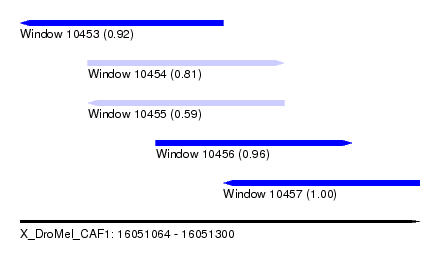
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 16,051,064 – 16,051,300 |
| Length | 236 |
| Max. P | 0.996049 |

| Location | 16,051,064 – 16,051,184 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.25 |
| Mean single sequence MFE | -52.90 |
| Consensus MFE | -49.74 |
| Energy contribution | -49.30 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.35 |
| Structure conservation index | 0.94 |
| SVM decision value | 1.12 |
| SVM RNA-class probability | 0.918499 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16051064 120 - 22224390 CGAAACGGUUUCGUCCACUCAGACGACAGUGUGCGAGCGGGCGCGAGAUAGCCGCACGAGCGUGCCAAGCCGCUCGCACGCACGGGACGGCGCUCUUUCGCGGCGUGAGUAAGAGAGUGG ((...((...(((((......)))))..((((((((((((((........)))((((....)))).....))))))))))).))...)).((((((((....((....)).)))))))). ( -53.40) >DroSec_CAF1 5259 120 - 1 CGAAACGGUUUCGUCCACUCAGACGACAGUGUGCGAGCGGGCGCGAGAUAGCCGCACGAGCGUGCCAAACCGCUCGCACGCACGGGACGGCGCUCUUUCGUGGCGAGAGUAAGAGAGUGG ((...((...(((((......)))))..((((((((((((((........)))((((....)))).....))))))))))).))...)).((((((((....((....)).)))))))). ( -52.40) >DroSim_CAF1 1952 120 - 1 CGAAACGGUUUCGUCCACUCAGACGACAGUGUGCGAGCGGGCGCGAGAUAGCCGCACGAGCGUGCCAAACCGCUCGCACGCACGGCACGGCGCUCUUUCGCGGCAGGAGUAAGAGAGUGG ((...((...(((((......)))))..((((((((((((((........)))((((....)))).....))))))))))).))...)).((((((((..(.......)..)))))))). ( -52.70) >DroEre_CAF1 5751 120 - 1 CGAAACGGUUUCGUCCACUCAGACGACAGUGUGCGAACGGGCGCGAGAUAGCCGCACGAGCGUGCCACACUGCUCGCACGCACUGCUCGGCGCUCUUUCGCGGGGAGAGUAAGAGAGCGG (((..((((.(((((......)))))..((((((((.(((((........)))((((....)))).....)).)))))))))))).))).((((((((..(.......)..)))))))). ( -47.40) >DroYak_CAF1 1663 120 - 1 CGAAACGGUUUCGUCCACUCAGACGACAGUGUGCGAGCGGGCGCGAGAUAGCCGCACGAGCGUGCCGCACCGCUCGCACGCACUGGACGGCGCUCUUUUGUGGCGAGAGUAAGAGAGCGG ((...((((.(((((......)))))..((((((((((((((((......)).((((....)))).)).))))))))))))))))..)).(((((((((...((....))))))))))). ( -58.60) >consensus CGAAACGGUUUCGUCCACUCAGACGACAGUGUGCGAGCGGGCGCGAGAUAGCCGCACGAGCGUGCCAAACCGCUCGCACGCACGGGACGGCGCUCUUUCGCGGCGAGAGUAAGAGAGUGG .....((...(((((......)))))..((((((((((((((........)))((((....)))).....)))))))))))......)).((((((((..(.......)..)))))))). (-49.74 = -49.30 + -0.44)


| Location | 16,051,104 – 16,051,220 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.77 |
| Mean single sequence MFE | -50.52 |
| Consensus MFE | -45.82 |
| Energy contribution | -45.86 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.37 |
| Structure conservation index | 0.91 |
| SVM decision value | 0.64 |
| SVM RNA-class probability | 0.810029 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16051104 116 + 22224390 CGUGCGAGCGGCUUGGCACGCUCGUGCGGCUAUCUCGCGCCCGCUCGCACACUGUCGUCUGAGUGGACGAAACCGUUUCGUUGGGUUGACACAUGCGACAUGCGACGUGUGU----GUGU ((..((((((((...)).))))))..)).......(((((.((((((((...((((((.((.((.(((..(((......)))..))).)).)).))))))))))).))).))----))). ( -50.40) >DroSec_CAF1 5299 116 + 1 CGUGCGAGCGGUUUGGCACGCUCGUGCGGCUAUCUCGCGCCCGCUCGCACACUGUCGUCUGAGUGGACGAAACCGUUUCGUUGGGUUGACACAUGCGACAUGCGACGUGUGU----GUGU ((..((((((........))))))..)).......(((((.((((((((...((((((.((.((.(((..(((......)))..))).)).)).))))))))))).))).))----))). ( -50.00) >DroSim_CAF1 1992 116 + 1 CGUGCGAGCGGUUUGGCACGCUCGUGCGGCUAUCUCGCGCCCGCUCGCACACUGUCGUCUGAGUGGACGAAACCGUUUCGUUGGGUUGACACAUGCGACAUGCGACGUGUGU----GUGU ((..((((((........))))))..)).......(((((.((((((((...((((((.((.((.(((..(((......)))..))).)).)).))))))))))).))).))----))). ( -50.00) >DroEre_CAF1 5791 120 + 1 CGUGCGAGCAGUGUGGCACGCUCGUGCGGCUAUCUCGCGCCCGUUCGCACACUGUCGUCUGAGUGGACGAAACCGUUUCGUUGGGUUGACACAUGCGACAUGCGACGUGUGUGUGUGUGU .(((((((....((.((((....)))).))...))))))).....(((((((..((((((....))))))..(((......)))....(((((((((.....)).)))))))))))))). ( -49.30) >DroYak_CAF1 1703 116 + 1 CGUGCGAGCGGUGCGGCACGCUCGUGCGGCUAUCUCGCGCCCGCUCGCACACUGUCGUCUGAGUGGACGAAACCGUUUCGUUGGGUUGACACAUGCGACAUGCGACGUGUGU----GUGU .((((((((((.((.((((....)))).((......))))))))))))))....((((((....))))))..(((......)))....(((((((((........)))))))----)).. ( -52.90) >consensus CGUGCGAGCGGUUUGGCACGCUCGUGCGGCUAUCUCGCGCCCGCUCGCACACUGUCGUCUGAGUGGACGAAACCGUUUCGUUGGGUUGACACAUGCGACAUGCGACGUGUGU____GUGU .(((((((((.....((((....))))(((........))))))))))))....((((((....))))))..(((......)))....(((((((((.....)).)))))))........ (-45.82 = -45.86 + 0.04)


| Location | 16,051,104 – 16,051,220 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.77 |
| Mean single sequence MFE | -41.44 |
| Consensus MFE | -40.52 |
| Energy contribution | -40.56 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.08 |
| Structure conservation index | 0.98 |
| SVM decision value | 0.11 |
| SVM RNA-class probability | 0.587311 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16051104 116 - 22224390 ACAC----ACACACGUCGCAUGUCGCAUGUGUCAACCCAACGAAACGGUUUCGUCCACUCAGACGACAGUGUGCGAGCGGGCGCGAGAUAGCCGCACGAGCGUGCCAAGCCGCUCGCACG ....----..((((((.((.....)))))))).((((.........))))(((((......)))))....((((((((((((........)))((((....)))).....))))))))). ( -42.10) >DroSec_CAF1 5299 116 - 1 ACAC----ACACACGUCGCAUGUCGCAUGUGUCAACCCAACGAAACGGUUUCGUCCACUCAGACGACAGUGUGCGAGCGGGCGCGAGAUAGCCGCACGAGCGUGCCAAACCGCUCGCACG ....----..((((((.((.....)))))))).((((.........))))(((((......)))))....((((((((((((........)))((((....)))).....))))))))). ( -42.10) >DroSim_CAF1 1992 116 - 1 ACAC----ACACACGUCGCAUGUCGCAUGUGUCAACCCAACGAAACGGUUUCGUCCACUCAGACGACAGUGUGCGAGCGGGCGCGAGAUAGCCGCACGAGCGUGCCAAACCGCUCGCACG ....----..((((((.((.....)))))))).((((.........))))(((((......)))))....((((((((((((........)))((((....)))).....))))))))). ( -42.10) >DroEre_CAF1 5791 120 - 1 ACACACACACACACGUCGCAUGUCGCAUGUGUCAACCCAACGAAACGGUUUCGUCCACUCAGACGACAGUGUGCGAACGGGCGCGAGAUAGCCGCACGAGCGUGCCACACUGCUCGCACG .............(((.((...(((((((((((((((.........))))..(((......))))))).)))))))....))(((((.(((..((((....))))....))))))))))) ( -35.90) >DroYak_CAF1 1703 116 - 1 ACAC----ACACACGUCGCAUGUCGCAUGUGUCAACCCAACGAAACGGUUUCGUCCACUCAGACGACAGUGUGCGAGCGGGCGCGAGAUAGCCGCACGAGCGUGCCGCACCGCUCGCACG ....----..((((((.((.....)))))))).((((.........))))(((((......)))))....((((((((((((((......)).((((....)))).)).)))))))))). ( -45.00) >consensus ACAC____ACACACGUCGCAUGUCGCAUGUGUCAACCCAACGAAACGGUUUCGUCCACUCAGACGACAGUGUGCGAGCGGGCGCGAGAUAGCCGCACGAGCGUGCCAAACCGCUCGCACG ..........((((((.((.....)))))))).((((.........))))(((((......)))))....((((((((((((........)))((((....)))).....))))))))). (-40.52 = -40.56 + 0.04)



| Location | 16,051,144 – 16,051,260 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 97.62 |
| Mean single sequence MFE | -46.29 |
| Consensus MFE | -43.76 |
| Energy contribution | -43.60 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.59 |
| Structure conservation index | 0.95 |
| SVM decision value | 1.54 |
| SVM RNA-class probability | 0.962728 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16051144 116 + 22224390 CCGCUCGCACACUGUCGUCUGAGUGGACGAAACCGUUUCGUUGGGUUGACACAUGCGACAUGCGACGUGUGU----GUGUGUGGUCAGUGCGGCACACACAUUCACACGCACUGCCGCCA ..(((((((...((((((.((.((.(((..(((......)))..))).)).)).)))))))))))((((((.----((((((((((.....))).))))))).)))))).......)).. ( -45.50) >DroSec_CAF1 5339 116 + 1 CCGCUCGCACACUGUCGUCUGAGUGGACGAAACCGUUUCGUUGGGUUGACACAUGCGACAUGCGACGUGUGU----GUGUGUGGUCAGUGCGGCACACACAUUCACACGCACUGCCGCCA ..(((((((...((((((.((.((.(((..(((......)))..))).)).)).)))))))))))((((((.----((((((((((.....))).))))))).)))))).......)).. ( -45.50) >DroSim_CAF1 2032 116 + 1 CCGCUCGCACACUGUCGUCUGAGUGGACGAAACCGUUUCGUUGGGUUGACACAUGCGACAUGCGACGUGUGU----GUGUGUGGUCAGUGCGGCACACACAUUCACACGCACUGCCGCCA ..(((((((...((((((.((.((.(((..(((......)))..))).)).)).)))))))))))((((((.----((((((((((.....))).))))))).)))))).......)).. ( -45.50) >DroEre_CAF1 5831 120 + 1 CCGUUCGCACACUGUCGUCUGAGUGGACGAAACCGUUUCGUUGGGUUGACACAUGCGACAUGCGACGUGUGUGUGUGUGUGUGGUCAGUGCGGCACACACAUUCACACGCACCGCCGCCA ..((.((((...((((((.((.((.(((..(((......)))..))).)).)).))))))))))))(((((((((.((((((((((.....))).))))))).)))))))))........ ( -50.40) >DroYak_CAF1 1743 116 + 1 CCGCUCGCACACUGUCGUCUGAGUGGACGAAACCGUUUCGUUGGGUUGACACAUGCGACAUGCGACGUGUGU----GUGUGUGGUCAGUGCGGCACACACAUUUACACGCACUGCCGCCA ..((.(((((((.((((((.((.(.((((((.....)))))).).)))))....((.....)))))))))))----).))((((.(((((((...............))))))))))).. ( -44.56) >consensus CCGCUCGCACACUGUCGUCUGAGUGGACGAAACCGUUUCGUUGGGUUGACACAUGCGACAUGCGACGUGUGU____GUGUGUGGUCAGUGCGGCACACACAUUCACACGCACUGCCGCCA ..(((((((...((((((.((.((.(((..(((......)))..))).)).)).))))))))))).((((((....((((((((((.....))).)))))))....))))))....)).. (-43.76 = -43.60 + -0.16)


| Location | 16,051,184 – 16,051,300 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.94 |
| Mean single sequence MFE | -49.82 |
| Consensus MFE | -43.72 |
| Energy contribution | -43.96 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.35 |
| Structure conservation index | 0.88 |
| SVM decision value | 2.65 |
| SVM RNA-class probability | 0.996049 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16051184 116 - 22224390 ACCAGCAAGCGGCGAGAGCGAGCGCUGCGUUCUCUUUGGCUGGCGGCAGUGCGUGUGAAUGUGUGUGCCGCACUGACCACACAC----ACACACGUCGCAUGUCGCAUGUGUCAACCCAA ....(((..((((.((((.(((((...))))).)))).))))(((((((((((((((..((((((((..........)))))))----))))))).))).)))))).))).......... ( -49.00) >DroSec_CAF1 5379 116 - 1 ACCAGCAAGAGGAGAGCGCGAGCGCUGCGUUCUCUUUGGCUGGCGGCAGUGCGUGUGAAUGUGUGUGCCGCACUGACCACACAC----ACACACGUCGCAUGUCGCAUGUGUCAACCCAA ..((((..((((((((((((.....)))))))))))).))))(((((((((((((((..((((((((..........)))))))----))))))).))).)))))).............. ( -55.10) >DroSim_CAF1 2072 116 - 1 ACCAGCAAGCGGCGAGAGCGAGCGCUGCGUUCUCUUUGGCUGGCGGCAGUGCGUGUGAAUGUGUGUGCCGCACUGACCACACAC----ACACACGUCGCAUGUCGCAUGUGUCAACCCAA ....(((..((((.((((.(((((...))))).)))).))))(((((((((((((((..((((((((..........)))))))----))))))).))).)))))).))).......... ( -49.00) >DroEre_CAF1 5871 120 - 1 ACCAGCAAGCGGCGAGAGCGAGCGCUGCGUUCUCUUUGGCUGGCGGCGGUGCGUGUGAAUGUGUGUGCCGCACUGACCACACACACACACACACGUCGCAUGUCGCAUGUGUCAACCCAA ....(((..((((.((((.(((((...))))).)))).))))(((((((((((((((..((((((((............))))))))..)))))).))).)))))).))).......... ( -49.90) >DroYak_CAF1 1783 116 - 1 ACCAGCAAGCGGCGAGAGCGAGCGCUGCGUUCUCUUUGGCUGGCGGCAGUGCGUGUAAAUGUGUGUGCCGCACUGACCACACAC----ACACACGUCGCAUGUCGCAUGUGUCAACCCAA ....(((..((((.((((.(((((...))))).)))).))))((((((((((((((...((((((((..........)))))))----).))))).))).)))))).))).......... ( -46.10) >consensus ACCAGCAAGCGGCGAGAGCGAGCGCUGCGUUCUCUUUGGCUGGCGGCAGUGCGUGUGAAUGUGUGUGCCGCACUGACCACACAC____ACACACGUCGCAUGUCGCAUGUGUCAACCCAA ..........((.(((((((.......))))))).(((((..(((((((((((((((...(((((((..........))))))).....)))))).))).))))))....))))).)).. (-43.72 = -43.96 + 0.24)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:12:54 2006