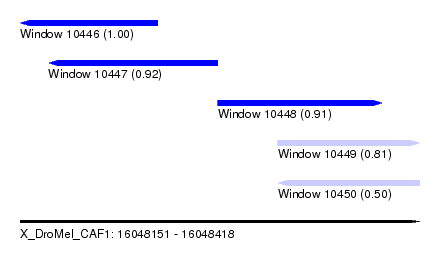
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 16,048,151 – 16,048,418 |
| Length | 267 |
| Max. P | 0.998843 |

| Location | 16,048,151 – 16,048,243 |
|---|---|
| Length | 92 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 70.89 |
| Mean single sequence MFE | -32.37 |
| Consensus MFE | -24.51 |
| Energy contribution | -24.73 |
| Covariance contribution | 0.23 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.80 |
| Structure conservation index | 0.76 |
| SVM decision value | 3.25 |
| SVM RNA-class probability | 0.998843 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16048151 92 - 22224390 AGGCAAUGUGCACUUACACAAGUAAUGCGCCAGUGUUAGUGUACAGUGGGCCUUGGCUACGACAGAAA---UUGGG-------------------UGCAUA------GGAACCAAAAUGA ((((..((((((((.((((..((.....))..)))).))))))))....))))...............---...((-------------------(.....------...)))....... ( -24.80) >DroSim_CAF1 93891 111 - 1 AGGCAAUGGGCACUUACACAAGUAAUGCGCCAGUGUGAGUGUACAGUGGGCCCUGGCUACGUCAGAAU---UUGGGAGAUCCAAACAAGACUAUUUGCAUA------GGAACCAAAAUGA ......((..(((((((((..((.....))..)))))))))..)).(((..((((((...(((....(---((((.....)))))...))).....)).))------))..)))...... ( -32.00) >DroYak_CAF1 98210 116 - 1 AGGCAAUGUGCACUUACACGCGCACUGCGCCAGUGUGAGUGUACAGUGGGUCUUGGCUACGCUGCACUUCUUCGGCAGAUCAAAGCAAAAG----UGCAAACAAACUGGAGUCAAAAUGU .(((..(((((((((((((((((...))))..)))))))))))))....)))((((((....(((((((.....((........))..)))----))))..(.....).))))))..... ( -40.30) >consensus AGGCAAUGUGCACUUACACAAGUAAUGCGCCAGUGUGAGUGUACAGUGGGCCUUGGCUACGACAGAAU___UUGGGAGAUC_AA_CAA_A_____UGCAUA______GGAACCAAAAUGA ..(((.(((((((((((((..((.....))..)))))))))))))((((.(....)))))...................................)))...................... (-24.51 = -24.73 + 0.23)



| Location | 16,048,170 – 16,048,283 |
|---|---|
| Length | 113 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 86.27 |
| Mean single sequence MFE | -37.10 |
| Consensus MFE | -31.68 |
| Energy contribution | -32.02 |
| Covariance contribution | 0.34 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.39 |
| Structure conservation index | 0.85 |
| SVM decision value | 1.11 |
| SVM RNA-class probability | 0.917016 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16048170 113 - 22224390 AGAACCGAAAAAGCUGUAAAAGAUUUCGCUCGAGUAAAUGAGGCAAUGUGCACUUACACAAGUAAUGCGCCAGUGUUAGUGUACAGUGGGCCUUGGCUACGACAGAAA---UUGGG---- ....((((.....((((........((....))(((..((((((..((((((((.((((..((.....))..)))).))))))))....))))))..))).))))...---)))).---- ( -33.80) >DroSim_CAF1 93925 117 - 1 AGAACCGAAAAAGCUGUAAAAGAUUUCGCUCAAGUAAAUGAGGCAAUGGGCACUUACACAAGUAAUGCGCCAGUGUGAGUGUACAGUGGGCCCUGGCUACGUCAGAAU---UUGGGAGAU .....(((((...((.....)).)))))(((((((......(((..((..(((((((((..((.....))..)))))))))..))....)))(((((...))))).))---))))).... ( -34.00) >DroYak_CAF1 98246 120 - 1 AGAACCGAAAAAGCUGUAAAAGAUUUCGCUCAAGUAAAUGAGGCAAUGUGCACUUACACGCGCACUGCGCCAGUGUGAGUGUACAGUGGGUCUUGGCUACGCUGCACUUCUUCGGCAGAU ....(((((.(((.((((...((......))..(((..((((((..(((((((((((((((((...))))..)))))))))))))....))))))..)))..))))))).)))))..... ( -43.50) >consensus AGAACCGAAAAAGCUGUAAAAGAUUUCGCUCAAGUAAAUGAGGCAAUGUGCACUUACACAAGUAAUGCGCCAGUGUGAGUGUACAGUGGGCCUUGGCUACGACAGAAU___UUGGGAGAU ....((((...(((.............)))...(((..((((((..(((((((((((((..((.....))..)))))))))))))....))))))..)))...........))))..... (-31.68 = -32.02 + 0.34)

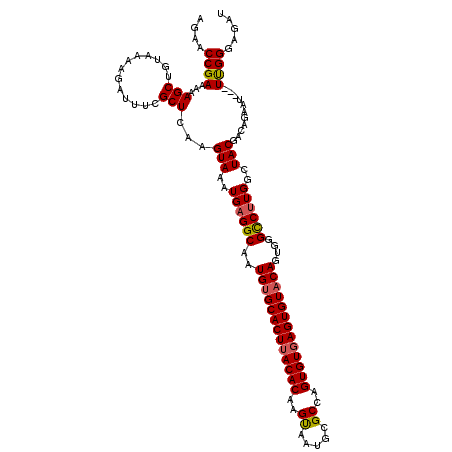
| Location | 16,048,283 – 16,048,393 |
|---|---|
| Length | 110 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.43 |
| Mean single sequence MFE | -38.73 |
| Consensus MFE | -36.97 |
| Energy contribution | -36.97 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.98 |
| Structure conservation index | 0.95 |
| SVM decision value | 1.10 |
| SVM RNA-class probability | 0.914613 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16048283 110 + 22224390 UCUGGCCAUUAAAAUCGCCUCUGACAACGUCGACGGCGACGACAACAUGCGGCGAUCAGUGUCGGACUUCGACUUUGGAUC----------CGAUCGGUCGGAGUUAAAGCCAAAACCAA ..((((((((...((((((.(((....(((((....)))))....)).).)))))).))))...(((((((((((((....----------)))..))))))))))...))))....... ( -36.60) >DroSim_CAF1 94042 106 + 1 UCUGGCCAUUAAAAUCGCCUCUGACAACGUCGACGGCGACGACAACAUGCGGCGAUCAGUGUCGGACUUCGACUUUGGAUC----------GGAUCGGUCGGAGUUAAAGCCAAAA---- ..((((((((...((((((.(((....(((((....)))))....)).).)))))).))))...((((((((((...(((.----------..)))))))))))))...))))...---- ( -34.00) >DroYak_CAF1 98366 120 + 1 UCUGGCCAUUAAAAUCGCCUCUGACAACGUCGACGGCGACGACAACAUGCGGCGAUCAGUGUCGGACUUCGACUUUGGAUCCGAUCCGAUCGGAUCGGUCGGAGUUAAAGCCAAAACCAA ..((((((((...((((((.(((....(((((....)))))....)).).)))))).))))...((((((((((...((((((((...))))))))))))))))))...))))....... ( -45.60) >consensus UCUGGCCAUUAAAAUCGCCUCUGACAACGUCGACGGCGACGACAACAUGCGGCGAUCAGUGUCGGACUUCGACUUUGGAUC__________GGAUCGGUCGGAGUUAAAGCCAAAACCAA ..((((((((...((((((.(((....(((((....)))))....)).).)))))).))))...((((((((((...((((...........))))))))))))))...))))....... (-36.97 = -36.97 + -0.00)



| Location | 16,048,323 – 16,048,418 |
|---|---|
| Length | 95 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 78.51 |
| Mean single sequence MFE | -27.67 |
| Consensus MFE | -21.37 |
| Energy contribution | -21.37 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.55 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.64 |
| SVM RNA-class probability | 0.808329 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16048323 95 + 22224390 GACAACAUGCGGCGAUCAGUGUCGGACUUCGACUUUGGAUC----------CGAUCGGUCGGAGUUAAAGCCAAAACCAAAACCAAAAGCAAAGCUAAAGCCAGA--------------- ........((((((((....))).(((((((((((((....----------)))..))))))))))...)))...............(((...)))...))....--------------- ( -22.30) >DroSim_CAF1 94082 89 + 1 GACAACAUGCGGCGAUCAGUGUCGGACUUCGACUUUGGAUC----------GGAUCGGUCGGAGUUAAAGCCAAAA------CCAAAAGCCAAGCUAAAGCCGCA--------------- .......((((((..........((.(((..((((((.(((----------(...))))))))))..)))))....------.....(((...)))...))))))--------------- ( -26.10) >DroYak_CAF1 98406 120 + 1 GACAACAUGCGGCGAUCAGUGUCGGACUUCGACUUUGGAUCCGAUCCGAUCGGAUCGGUCGGAGUUAAAGCCAAAACCAAAACCAAAACCAAAGCUAAAGCCAGAGCCAGAGUCAAAAGA (((.......((((((....))).((((((((((...((((((((...))))))))))))))))))...))).....................(((........)))....)))...... ( -34.60) >consensus GACAACAUGCGGCGAUCAGUGUCGGACUUCGACUUUGGAUC__________GGAUCGGUCGGAGUUAAAGCCAAAACCAAAACCAAAAGCAAAGCUAAAGCCAGA_______________ ..........((((((....))).((((((((((...((((...........))))))))))))))...))).....................((....))................... (-21.37 = -21.37 + -0.00)



| Location | 16,048,323 – 16,048,418 |
|---|---|
| Length | 95 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 78.51 |
| Mean single sequence MFE | -31.50 |
| Consensus MFE | -18.80 |
| Energy contribution | -20.47 |
| Covariance contribution | 1.67 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.75 |
| Structure conservation index | 0.60 |
| SVM decision value | -0.06 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16048323 95 - 22224390 ---------------UCUGGCUUUAGCUUUGCUUUUGGUUUUGGUUUUGGCUUUAACUCCGACCGAUCG----------GAUCCAAAGUCGAAGUCCGACACUGAUCGCCGCAUGUUGUC ---------------...(((..((((...((..(..((.((((((((((((((...(((((....)))----------))...)))))))))).)))).))..)..)).)).))..))) ( -28.70) >DroSim_CAF1 94082 89 - 1 ---------------UGCGGCUUUAGCUUGGCUUUUGG------UUUUGGCUUUAACUCCGACCGAUCC----------GAUCCAAAGUCGAAGUCCGACACUGAUCGCCGCAUGUUGUC ---------------((((((........((...((((------((..((......))..)))))).))----------((((....((((.....))))...))))))))))....... ( -26.80) >DroYak_CAF1 98406 120 - 1 UCUUUUGACUCUGGCUCUGGCUUUAGCUUUGGUUUUGGUUUUGGUUUUGGCUUUAACUCCGACCGAUCCGAUCGGAUCGGAUCCAAAGUCGAAGUCCGACACUGAUCGCCGCAUGUUGUC ......(((....((...(((.((((..(((((((((..(((((..((((........))))(((((((....)))))))..)))))..))))).))))..))))..))))).....))) ( -39.00) >consensus _______________UCUGGCUUUAGCUUUGCUUUUGGUUUUGGUUUUGGCUUUAACUCCGACCGAUCC__________GAUCCAAAGUCGAAGUCCGACACUGAUCGCCGCAUGUUGUC ..................(((((..((((((.....((((..((((........))))..))))((((...........))))))))))..)))))(((((.((.......))))))).. (-18.80 = -20.47 + 1.67)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:12:47 2006