
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 16,040,843 – 16,041,070 |
| Length | 227 |
| Max. P | 0.992380 |

| Location | 16,040,843 – 16,040,962 |
|---|---|
| Length | 119 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 78.59 |
| Mean single sequence MFE | -31.70 |
| Consensus MFE | -24.37 |
| Energy contribution | -23.60 |
| Covariance contribution | -0.77 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.67 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.86 |
| SVM RNA-class probability | 0.869155 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
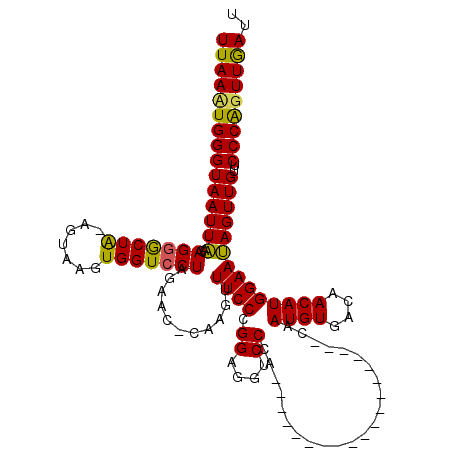
>X_DroMel_CAF1 16040843 119 + 22224390 UUAAAUGGGUAAUUAACAGGGCUA-AAUAAGUGGUCCUAGAACCCAAGUUUCCCGGAGGUCCCAAGUACCCCAGUGGCUUGGCCAAUGUGACAACAUGGAAUAGUUGUUCCCAGUUGAUU ((((.((((....(((((((((((-......))))))).....((((((..(..((.(((.......))))).)..))))))(((.(((....))))))....))))..)))).)))).. ( -33.20) >DroEre_CAF1 107785 98 + 1 UUAAUUGGGUAAUUGGGAGCGCUA-AGUAAGUGGUCCUAGAAC-CAGGUUUCCCGGAGGUCCC--------------------CAAUGUGACAACAUGGAACAGUUGUUCCCGGUUGAUU ...((((((...((((((.((((.-....)))).)))))).((-(.((....))...))).))--------------------))))....((((..(((((....)))))..))))... ( -32.40) >DroYak_CAF1 90783 102 + 1 UUAACUGGGUAAUUAAGAGGACUGGAGUAAGUGGUCCUAGAACUCAAGUUUCCCGGAGGUCCUAG------------------CAAUGUGACAACAUGGAAUAGUUGUUCCCAGUUAAUU (((((((((((((((.(((..(((((.(....).))).))..)))....((((..(..(((.((.------------------...)).)))..)..))))))))))..))))))))).. ( -29.50) >consensus UUAAAUGGGUAAUUAAGAGGGCUA_AGUAAGUGGUCCUAGAAC_CAAGUUUCCCGGAGGUCCCA___________________CAAUGUGACAACAUGGAAUAGUUGUUCCCAGUUGAUU (((((((((((((((..(((((((.......)))))))...........((((.((....)).......................((((....))))))))))))))..))))))))).. (-24.37 = -23.60 + -0.77)


| Location | 16,040,843 – 16,040,962 |
|---|---|
| Length | 119 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 78.59 |
| Mean single sequence MFE | -32.03 |
| Consensus MFE | -18.26 |
| Energy contribution | -18.60 |
| Covariance contribution | 0.34 |
| Combinations/Pair | 1.21 |
| Mean z-score | -3.32 |
| Structure conservation index | 0.57 |
| SVM decision value | 2.32 |
| SVM RNA-class probability | 0.992380 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16040843 119 - 22224390 AAUCAACUGGGAACAACUAUUCCAUGUUGUCACAUUGGCCAAGCCACUGGGGUACUUGGGACCUCCGGGAAACUUGGGUUCUAGGACCACUUAUU-UAGCCCUGUUAAUUACCCAUUUAA ...(((((((((.......))))).))))...((.((((...)))).))(((((((((((((((..((....)).)))))))))).........(-((((...))))).)))))...... ( -35.40) >DroEre_CAF1 107785 98 - 1 AAUCAACCGGGAACAACUGUUCCAUGUUGUCACAUUG--------------------GGGACCUCCGGGAAACCUG-GUUCUAGGACCACUUACU-UAGCGCUCCCAAUUACCCAAUUAA ...((((..(((((....)))))..))))....((((--------------------((((((...((....)).)-)))...(((.(.((....-.)).).)))......))))))... ( -28.60) >DroYak_CAF1 90783 102 - 1 AAUUAACUGGGAACAACUAUUCCAUGUUGUCACAUUG------------------CUAGGACCUCCGGGAAACUUGAGUUCUAGGACCACUUACUCCAGUCCUCUUAAUUACCCAGUUAA ..(((((((((.(((((........)))))...((((------------------..((((((((.((....)).))).....(((........))).)))))..))))..))))))))) ( -32.10) >consensus AAUCAACUGGGAACAACUAUUCCAUGUUGUCACAUUG___________________UGGGACCUCCGGGAAACUUG_GUUCUAGGACCACUUACU_UAGCCCUCUUAAUUACCCAAUUAA ........(((.(((((........))))).........................(((((((.((.((....)).)))))))))...........................)))...... (-18.26 = -18.60 + 0.34)



| Location | 16,040,882 – 16,041,002 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 76.90 |
| Mean single sequence MFE | -35.97 |
| Consensus MFE | -24.23 |
| Energy contribution | -24.47 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.22 |
| Mean z-score | -1.75 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.92 |
| SVM RNA-class probability | 0.881605 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16040882 120 + 22224390 AACCCAAGUUUCCCGGAGGUCCCAAGUACCCCAGUGGCUUGGCCAAUGUGACAACAUGGAAUAGUUGUUCCCAGUUGAUUGGGUUACUGCCUCCGGCGGCAACUUCAUGUUCUUAUGUGA .....(((((((((((((((....((((.(((((((((...))).......((((..(((((....)))))..)))))))))).)))))))))))).)).)))))(((((....))))). ( -41.90) >DroEre_CAF1 107824 97 + 1 AAC-CAGGUUUCCCGGAGGUCCC--------------------CAAUGUGACAACAUGGAACAGUUGUUCCCGGUUGAUUGGCACACUGCCUCC--GGGCAGCUCCGUGUUCCUAUGUGA .((-..((((.(((((((((..(--------------------((((....((((..(((((....)))))..)))))))))......))))))--))).))))..))............ ( -34.80) >DroYak_CAF1 90823 102 + 1 AACUCAAGUUUCCCGGAGGUCCUAG------------------CAAUGUGACAACAUGGAAUAGUUGUUCCCAGUUAAUUGGGAUACUGCCUCCAGGGGCAACUUCGUGUGCUCAUGUGA .....(((((((((((((((((...------------------..((((....))))))).((((...((((((....)))))).))))))))).)))).)))))((((....))))... ( -31.20) >consensus AAC_CAAGUUUCCCGGAGGUCCCA___________________CAAUGUGACAACAUGGAAUAGUUGUUCCCAGUUGAUUGGGAUACUGCCUCC_GGGGCAACUUCGUGUUCUUAUGUGA .....(((((.((((((((((((((....................((((....))))(((((....))))).......))))).....))))))..))).)))))((((....))))... (-24.23 = -24.47 + 0.24)



| Location | 16,040,882 – 16,041,002 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 76.90 |
| Mean single sequence MFE | -35.33 |
| Consensus MFE | -20.50 |
| Energy contribution | -19.73 |
| Covariance contribution | -0.77 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.88 |
| Structure conservation index | 0.58 |
| SVM decision value | 0.28 |
| SVM RNA-class probability | 0.665356 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16040882 120 - 22224390 UCACAUAAGAACAUGAAGUUGCCGCCGGAGGCAGUAACCCAAUCAACUGGGAACAACUAUUCCAUGUUGUCACAUUGGCCAAGCCACUGGGGUACUUGGGACCUCCGGGAAACUUGGGUU .........(((.(.(((((..(.((((((((((((.(((...(((((((((.......))))).))))...((.((((...)))).)))))))))...).)))))))).))))).)))) ( -38.40) >DroEre_CAF1 107824 97 - 1 UCACAUAGGAACACGGAGCUGCCC--GGAGGCAGUGUGCCAAUCAACCGGGAACAACUGUUCCAUGUUGUCACAUUG--------------------GGGACCUCCGGGAAACCUG-GUU .........(((.(((.(...(((--((((((((((((.....((((..(((((....)))))..)))).)))))))--------------------....))))))))...))))-))) ( -37.20) >DroYak_CAF1 90823 102 - 1 UCACAUGAGCACACGAAGUUGCCCCUGGAGGCAGUAUCCCAAUUAACUGGGAACAACUAUUCCAUGUUGUCACAUUG------------------CUAGGACCUCCGGGAAACUUGAGUU ...............(((((...((((((((((((((((((......)))))(((((........))))).....))------------------))..).)))))))).)))))..... ( -30.40) >consensus UCACAUAAGAACACGAAGUUGCCCC_GGAGGCAGUAUCCCAAUCAACUGGGAACAACUAUUCCAUGUUGUCACAUUG___________________UGGGACCUCCGGGAAACUUG_GUU ..((.((((.........(((((......)))))...(((...((((..((((......))))..)))).............................((....)))))...)))).)). (-20.50 = -19.73 + -0.77)



| Location | 16,040,962 – 16,041,070 |
|---|---|
| Length | 108 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 80.46 |
| Mean single sequence MFE | -25.07 |
| Consensus MFE | -17.81 |
| Energy contribution | -17.23 |
| Covariance contribution | -0.58 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.59 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.16 |
| SVM RNA-class probability | 0.613943 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16040962 108 - 22224390 CAUUUUAAUUUACAAUUCAUGGAAACAUGAUGGGUUGUCUUCGCACACAACCACAUUUGCAUUC------------ACAUUCACAUAAGAACAUGAAGUUGCCGCCGGAGGCAGUAACCC ................(((((....)))))((((((((........)))))).))......(((------------(..(((......)))..))))(((((.(((...))).))))).. ( -26.30) >DroEre_CAF1 107883 105 - 1 CAUUUUAAUUUACGUUUCAUGGAAAAAUGAUGGCUUGUCUUC-CACACAGCCACAUUAACAUUC------------ACAUUCACAUAGGAACACGGAGCUGCCC--GGAGGCAGUGUGCC .............(((((((((((.((((.((((((((....-.))).))))))))).......------------...))).))).)))))..((.((((((.--...))))))...)) ( -25.70) >DroYak_CAF1 90885 120 - 1 CAUUUUAAUUUACAUUUCAUGGAAACAUGAUGGGUUGUCUUCGCACACAACCACACUCACAUUCACUAUAACCAUCACAUUCACAUGAGCACACGAAGUUGCCCCUGGAGGCAGUAUCCC ...............(((((((((...(((((((((((........))))))........((.....))...)))))..))).)))))).....((..(((((......)))))..)).. ( -23.20) >consensus CAUUUUAAUUUACAUUUCAUGGAAACAUGAUGGGUUGUCUUCGCACACAACCACAUUAACAUUC____________ACAUUCACAUAAGAACACGAAGUUGCCCC_GGAGGCAGUAUCCC ................(((((....))))).((((((((((((....((((..(........................................)..))))....)))))))))...))) (-17.81 = -17.23 + -0.58)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:12:40 2006