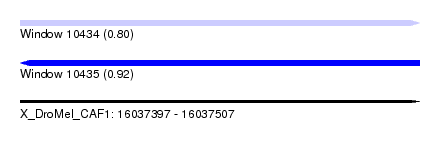
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 16,037,397 – 16,037,507 |
| Length | 110 |
| Max. P | 0.915042 |

| Location | 16,037,397 – 16,037,507 |
|---|---|
| Length | 110 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 80.06 |
| Mean single sequence MFE | -33.70 |
| Consensus MFE | -21.47 |
| Energy contribution | -21.80 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.02 |
| Structure conservation index | 0.64 |
| SVM decision value | 0.61 |
| SVM RNA-class probability | 0.798605 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16037397 110 + 22224390 AUGUGUUAAUUGCACUUGGCAGUGUUUUUCCUGAAAAGUGAUCCUUUUGGCCUCUC-----GGCCAUUUGUCCCUUUGGCCAGAUGAUGUAUAGCAGAUAAAACGGCACUGCCGC----- ..((((.....))))..(((((((((((..(((....((((((..(((((((....-----((.(....).))....))))))).))).)))..)))..)))...))))))))..----- ( -33.60) >DroEre_CAF1 104976 97 + 1 AUGUGUUAAUUGCACUUGGCAGUGUUUUUCCUGAAAAGCGAUCCUUUUGGC---------------------CCUUUGGCCAGAUGAGGUCU--CAGAUAAAACGCCACCGCCGCCAACA ..((((.....))))..(((.(((((((..((((.....(((((.((((((---------------------(....))))))).).)))))--)))..)))))))....)))....... ( -31.10) >DroYak_CAF1 87717 116 + 1 AUGUGUUAAUUGCACUUGGCUGUGUUUUUCCUGAAAAGUGAUCCUUUUGGCCUCUCCGGCCAGCCAUUUGUCCCUUUGGCCAGAUGAGGUAU-CCAGAUAAAACGGCACUGCCGCCA--- ..((((.....)))).((((...(((((..((((((((.....))))).(((((((.((((((............)))))).)).)))))..-.)))..)))))(((...)))))))--- ( -36.40) >consensus AUGUGUUAAUUGCACUUGGCAGUGUUUUUCCUGAAAAGUGAUCCUUUUGGCCUCUC______GCCAUUUGUCCCUUUGGCCAGAUGAGGUAU__CAGAUAAAACGGCACUGCCGCCA___ ..((((.....))))..(((.(((((((..(((........((..(((((((.........................))))))).)).......)))..)))))...)).)))....... (-21.47 = -21.80 + 0.33)


| Location | 16,037,397 – 16,037,507 |
|---|---|
| Length | 110 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 80.06 |
| Mean single sequence MFE | -33.95 |
| Consensus MFE | -21.72 |
| Energy contribution | -22.28 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.28 |
| Structure conservation index | 0.64 |
| SVM decision value | 1.10 |
| SVM RNA-class probability | 0.915042 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16037397 110 - 22224390 -----GCGGCAGUGCCGUUUUAUCUGCUAUACAUCAUCUGGCCAAAGGGACAAAUGGCC-----GAGAGGCCAAAAGGAUCACUUUUCAGGAAAAACACUGCCAAGUGCAAUUAACACAU -----..(((((((...((((.((((......(((...((.((....)).))..(((((-----....)))))....))).......)))))))).)))))))..(((.......))).. ( -31.32) >DroEre_CAF1 104976 97 - 1 UGUUGGCGGCGGUGGCGUUUUAUCUG--AGACCUCAUCUGGCCAAAGG---------------------GCCAAAAGGAUCGCUUUUCAGGAAAAACACUGCCAAGUGCAAUUAACACAU ((((((..(((.(((((((((.((((--((((((....(((((....)---------------------))))..)))......))))))).)))))...))))..)))..))))))... ( -31.20) >DroYak_CAF1 87717 116 - 1 ---UGGCGGCAGUGCCGUUUUAUCUGG-AUACCUCAUCUGGCCAAAGGGACAAAUGGCUGGCCGGAGAGGCCAAAAGGAUCACUUUUCAGGAAAAACACAGCCAAGUGCAAUUAACACAU ---(((((((...)))(((((.(((((-(..((((.((((((((..............))))))))))))((....)).......)))))).)))))...)))).(((.......))).. ( -39.34) >consensus ___UGGCGGCAGUGCCGUUUUAUCUG__AUACCUCAUCUGGCCAAAGGGACAAAUGGC______GAGAGGCCAAAAGGAUCACUUUUCAGGAAAAACACUGCCAAGUGCAAUUAACACAU .......(((((((...((((.((((.............((((.........................))))(((((.....))))))))))))).)))))))..(((.......))).. (-21.72 = -22.28 + 0.56)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:12:33 2006