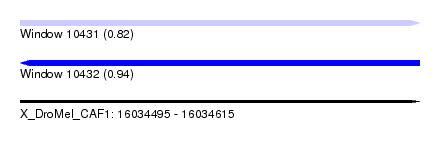
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 16,034,495 – 16,034,615 |
| Length | 120 |
| Max. P | 0.940091 |

| Location | 16,034,495 – 16,034,615 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 80.62 |
| Mean single sequence MFE | -37.90 |
| Consensus MFE | -24.84 |
| Energy contribution | -25.07 |
| Covariance contribution | 0.23 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.05 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.67 |
| SVM RNA-class probability | 0.819263 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16034495 120 + 22224390 CAAUGGAUGAUGGGAUGCUGGGGAUGCUGCGAUGCUGGGCAGGAUGCCCCAGGAUUUCCGGCAAUCCUUUUGUAAGGUUGGGGUAAUAGGGUGGUUAACGCCACAUAUACAAAUAGGCAU ....((((..((((((.((((((((.((((........)))).)).)))))).).)))))...))))...(((...(((...(((.((..(((((....))))).))))).)))..))). ( -40.70) >DroEre_CAF1 102253 100 + 1 AUGUGGAUGUUGGGUU-----------------GCCGGGCAGGAUGCCCCAGGAUUUCCGGCAAUCCUCCGG---AGGAGGGAUAAUAGGGUGGUUAACGCCACAUAUACAAAUAGGCAU ((((((.(((((((((-----------------(((((((.....))))..((....))))))))(((((..---.))))).............)))))))))))).............. ( -35.90) >DroYak_CAF1 85013 116 + 1 AAAUGGAUGUUGGGAUGUUG-GGAUGCUGGGAAGCUGGGCAGGAUGCCCCAGGAUUUCCGGCAAUCCUUUGG---AGGAGGGAUAAUAGGGUGGUUAACGCCACAUAUACAAAUAGGCAU ..(((..((((.(.((((..-...(((((((((.((((((.....).)))))..)))))))))(((((((..---..))))))).....((((.....))))))))...).))))..))) ( -37.10) >consensus AAAUGGAUGUUGGGAUG_UG_GGAUGCUG_GA_GCUGGGCAGGAUGCCCCAGGAUUUCCGGCAAUCCUUUGG___AGGAGGGAUAAUAGGGUGGUUAACGCCACAUAUACAAAUAGGCAU ..(((..((((.(....................(((((((.....))))..((....)))))..((((((......))))))........(((((....))))).....).))))..))) (-24.84 = -25.07 + 0.23)

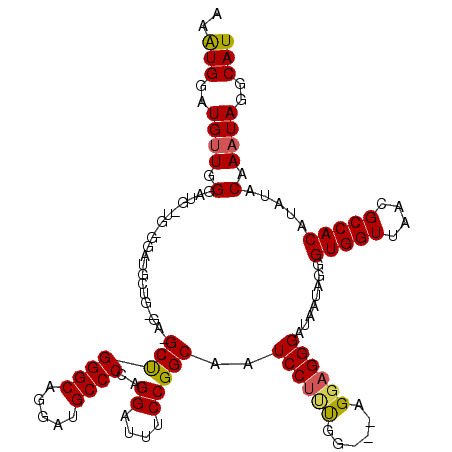
| Location | 16,034,495 – 16,034,615 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 80.62 |
| Mean single sequence MFE | -27.97 |
| Consensus MFE | -19.13 |
| Energy contribution | -19.13 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.30 |
| Structure conservation index | 0.68 |
| SVM decision value | 1.28 |
| SVM RNA-class probability | 0.940091 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16034495 120 - 22224390 AUGCCUAUUUGUAUAUGUGGCGUUAACCACCCUAUUACCCCAACCUUACAAAAGGAUUGCCGGAAAUCCUGGGGCAUCCUGCCCAGCAUCGCAGCAUCCCCAGCAUCCCAUCAUCCAUUG .......((((((....(((.((.............)).)))....)))))).((((....(((....((((((.((.((((........)))).))))))))..)))....)))).... ( -30.52) >DroEre_CAF1 102253 100 - 1 AUGCCUAUUUGUAUAUGUGGCGUUAACCACCCUAUUAUCCCUCCU---CCGGAGGAUUGCCGGAAAUCCUGGGGCAUCCUGCCCGGC-----------------AACCCAACAUCCACAU ((((......))))((((((.(((...............(((((.---..))))).(((((((....))..((((.....)))))))-----------------))...)))..)))))) ( -30.90) >DroYak_CAF1 85013 116 - 1 AUGCCUAUUUGUAUAUGUGGCGUUAACCACCCUAUUAUCCCUCCU---CCAAAGGAUUGCCGGAAAUCCUGGGGCAUCCUGCCCAGCUUCCCAGCAUCC-CAACAUCCCAACAUCCAUUU ((((((((......))).)))))......................---.....((..(((.((((...(((((........))))).))))..)))..)-)................... ( -22.50) >consensus AUGCCUAUUUGUAUAUGUGGCGUUAACCACCCUAUUAUCCCUCCU___CCAAAGGAUUGCCGGAAAUCCUGGGGCAUCCUGCCCAGC_UC_CAGCAUCC_CA_CAUCCCAACAUCCAUUU ((((((((......))).))))).............................((((((......)))))).((((.....)))).................................... (-19.13 = -19.13 + 0.00)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:12:31 2006