
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 16,032,201 – 16,032,353 |
| Length | 152 |
| Max. P | 0.954177 |

| Location | 16,032,201 – 16,032,313 |
|---|---|
| Length | 112 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 61.60 |
| Mean single sequence MFE | -32.33 |
| Consensus MFE | -13.54 |
| Energy contribution | -12.89 |
| Covariance contribution | -0.65 |
| Combinations/Pair | 1.35 |
| Mean z-score | -1.96 |
| Structure conservation index | 0.42 |
| SVM decision value | 0.19 |
| SVM RNA-class probability | 0.626477 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16032201 112 + 22224390 AUUUCGUAUGUAACUUGAGUUCAAAGUCCCGAUUGUGUUUG------UCCAGAAACG--CAAGUGGCAAAAAAUCGGGCGCCAUGGACUCGUUAUCCUGCGCCCGCCCUUCCUAUGCCCA ....((((.(((((..(((((((..(((((((((...((((------((.....((.--...)))))))).))))))).))..))))))))))))..))))................... ( -32.30) >DroEre_CAF1 99995 101 + 1 AUUUCGUACGUUACUCAAGCUAAAAAGUUCGAUUGUACC-------------AAGUGGCAAAGUGGAA-AAAGUUGGGCGCCAGGAACUCGCCAUCCUGCGCU----CCUCCUUUGCC-C ((((.(((((....((.((((....)))).)).))))).-------------))))(((((((..(..-......((((((.((((........)))))))))----))..)))))))-. ( -30.50) >DroYak_CAF1 82560 111 + 1 A----AUGCGCUCUAUAAGCAUUGAAGUUCGAUUGCAUUUACAUAUACGCAGAAACCAAAAAGUGGCA-AAAGUCGGGCGCCAGGAACUCGUAAUCCUGCGCU----CCUCCUUUGCCAC (----(((((((.....)))((((.....)))).))))).......................((((((-((.(..((((((.((((........)))))))))----)..).)))))))) ( -34.20) >consensus AUUUCGUACGUUACUUAAGCUAAAAAGUUCGAUUGUAUUU_________CAGAAACG__AAAGUGGCA_AAAGUCGGGCGCCAGGAACUCGUAAUCCUGCGCU____CCUCCUUUGCC_C ..................((......((((((((.....................................))))))))(((((((........))))).)).............))... (-13.54 = -12.89 + -0.65)


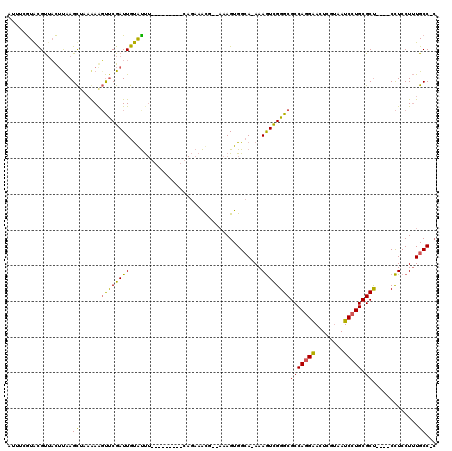
| Location | 16,032,241 – 16,032,353 |
|---|---|
| Length | 112 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 77.36 |
| Mean single sequence MFE | -34.32 |
| Consensus MFE | -20.34 |
| Energy contribution | -20.23 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.36 |
| Structure conservation index | 0.59 |
| SVM decision value | 1.44 |
| SVM RNA-class probability | 0.954177 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16032241 112 + 22224390 G------UCCAGAAACG--CAAGUGGCAAAAAAUCGGGCGCCAUGGACUCGUUAUCCUGCGCCCGCCCUUCCUAUGCCCAGAUAAUCGAUGCAUAAUAAAUGGCAUCACUGUCCCAUCUG .------..((((.((.--...))((((......(((((((...(((.......))).))))))).........))))..((((...(((((..........)))))..))))...)))) ( -32.06) >DroEre_CAF1 100034 102 + 1 ------------AAGUGGCAAAGUGGAA-AAAGUUGGGCGCCAGGAACUCGCCAUCCUGCGCU----CCUCCUUUGCC-CGAUAAUCGAUGCAUAAUAAAUGGCAUCACUGUCCCAUCGG ------------..((((...(((((..-((((..((((((.((((........)))))))).----))..))))..)-).......(((((..........))))))))...))))... ( -32.00) >DroYak_CAF1 82596 115 + 1 ACAUAUACGCAGAAACCAAAAAGUGGCA-AAAGUCGGGCGCCAGGAACUCGUAAUCCUGCGCU----CCUCCUUUGCCACGAUAAUCGAUGCAUAAUAAAUGGCAUCACUGUCCCAUCGG ........((((..........((((((-((.(..((((((.((((........)))))))))----)..).)))))))).......(((((..........))))).))))........ ( -38.90) >consensus _________CAGAAACG__AAAGUGGCA_AAAGUCGGGCGCCAGGAACUCGUAAUCCUGCGCU____CCUCCUUUGCC_CGAUAAUCGAUGCAUAAUAAAUGGCAUCACUGUCCCAUCGG ......................((((..........(((((.((((........))))))))).................((((...(((((..........)))))..))))))))... (-20.34 = -20.23 + -0.11)

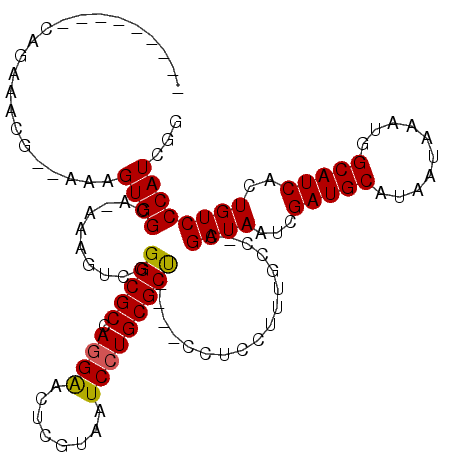
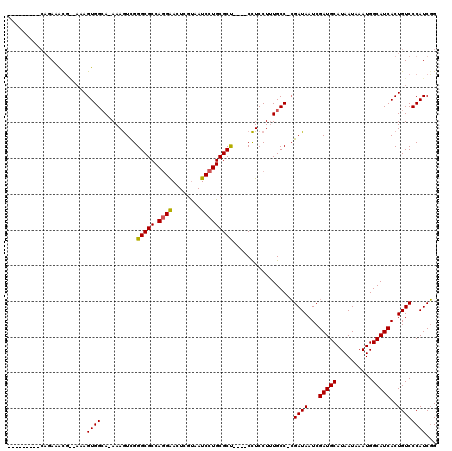
| Location | 16,032,241 – 16,032,353 |
|---|---|
| Length | 112 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 77.36 |
| Mean single sequence MFE | -39.60 |
| Consensus MFE | -24.80 |
| Energy contribution | -24.03 |
| Covariance contribution | -0.77 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.13 |
| Structure conservation index | 0.63 |
| SVM decision value | 1.34 |
| SVM RNA-class probability | 0.943944 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16032241 112 - 22224390 CAGAUGGGACAGUGAUGCCAUUUAUUAUGCAUCGAUUAUCUGGGCAUAGGAAGGGCGGGCGCAGGAUAACGAGUCCAUGGCGCCCGAUUUUUUGCCACUUG--CGUUUCUGGA------C (((((((.....((((((.((.....)))))))).)))))))((((.(((((...(((((((.((((.....))))...))))))).))))))))).....--..........------. ( -40.60) >DroEre_CAF1 100034 102 - 1 CCGAUGGGACAGUGAUGCCAUUUAUUAUGCAUCGAUUAUCG-GGCAAAGGAGG----AGCGCAGGAUGGCGAGUUCCUGGCGCCCAACUUU-UUCCACUUUGCCACUU------------ .((((((.....((((((.((.....)))))))).))))))-(((((((..((----(((((((((........)))).))))........-.))).)))))))....------------ ( -35.60) >DroYak_CAF1 82596 115 - 1 CCGAUGGGACAGUGAUGCCAUUUAUUAUGCAUCGAUUAUCGUGGCAAAGGAGG----AGCGCAGGAUUACGAGUUCCUGGCGCCCGACUUU-UGCCACUUUUUGGUUUCUGCGUAUAUGU ..(((((..((....)))))))...((((((..(((((..((((((((((.((----.((((((((........)))).))))))..))))-))))))....)))))..))))))..... ( -42.60) >consensus CCGAUGGGACAGUGAUGCCAUUUAUUAUGCAUCGAUUAUCG_GGCAAAGGAGG____AGCGCAGGAUAACGAGUUCCUGGCGCCCGACUUU_UGCCACUUU__CGUUUCUG_________ .((((((.....((((((.((.....)))))))).)))))).((((...((((.....((((.((((.....))))...))))....)))).))))........................ (-24.80 = -24.03 + -0.77)

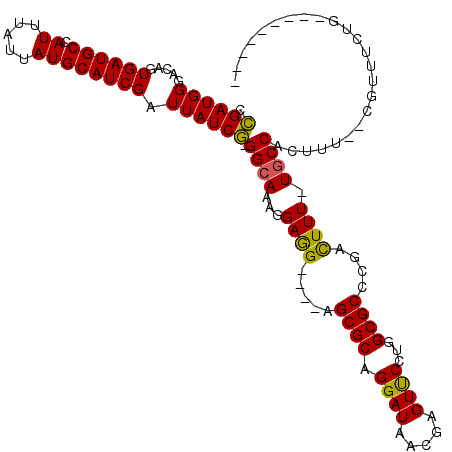
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:12:26 2006