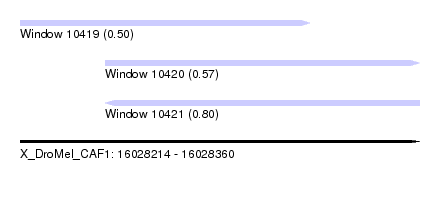
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 16,028,214 – 16,028,360 |
| Length | 146 |
| Max. P | 0.799120 |

| Location | 16,028,214 – 16,028,320 |
|---|---|
| Length | 106 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 80.22 |
| Mean single sequence MFE | -33.10 |
| Consensus MFE | -20.19 |
| Energy contribution | -19.97 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.79 |
| Structure conservation index | 0.61 |
| SVM decision value | -0.07 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16028214 106 + 22224390 AUCGGCAUCCC------CAUCUGCAACUCGUUUAU---GU----ACAUACAUAUAUUUAACGUAUAUUUUUGAGCUUGCGAA-AAAGGGGUGGGCUGCGGUUGGACCCGUGGCCCAUGUU ...((((.(((------(...(((((((((...((---((----((...............))))))...)))).)))))..-...))))(((((..(((......)))..))))))))) ( -37.06) >DroEre_CAF1 96427 113 + 1 AUCGGCAUCGCCAUUCCCAUCUGCAACUCCUUUUU-ACAUAUACACA----UAGAUUCAACGUAUAUUUUGGAGCUUCUGCCG--AAGGGUGGGCUGCGGUUGGACCCGUGGCCCAUGUU ...(((...)))...(((.((.(((.((((.....-..((((((...----..........))))))...))))....))).)--).)))(((((..(((......)))..))))).... ( -35.82) >DroYak_CAF1 78984 120 + 1 AUCGCCAUCGCCAUCCCCAUCUGCAACUCCUUUAUCUCAUAUACACAUACAUAUAUUUAACGUAUAUUUUUGAGCUUCUGGUAAAAAGGGUGGGCUGCGGUUGGACCCGUGGCCCAUGUU ...((((..((...........))...........(((((((((.................)))))....))))....))))....((.((((((..(((......)))..)))))).)) ( -26.43) >consensus AUCGGCAUCGCCAU_CCCAUCUGCAACUCCUUUAU__CAUAUACACAUACAUAUAUUUAACGUAUAUUUUUGAGCUUCUGAA_AAAAGGGUGGGCUGCGGUUGGACCCGUGGCCCAUGUU ......................................................................................((.((((((..(((......)))..)))))).)) (-20.19 = -19.97 + -0.22)



| Location | 16,028,245 – 16,028,360 |
|---|---|
| Length | 115 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 86.94 |
| Mean single sequence MFE | -33.87 |
| Consensus MFE | -27.61 |
| Energy contribution | -27.39 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.78 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.07 |
| SVM RNA-class probability | 0.566593 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16028245 115 + 22224390 ----ACAUACAUAUAUUUAACGUAUAUUUUUGAGCUUGCGAA-AAAGGGGUGGGCUGCGGUUGGACCCGUGGCCCAUGUUGUUGUGGUCACUUUUUCCACAUUGUUUUCCUUUUGCCAAU ----.((...((((((.....))))))...))...(((.(.(-((((((((((((..(((......)))..)))))).....(((((.........)))))......))))))).)))). ( -33.70) >DroEre_CAF1 96466 114 + 1 AUACACA----UAGAUUCAACGUAUAUUUUGGAGCUUCUGCCG--AAGGGUGGGCUGCGGUUGGACCCGUGGCCCAUGUUGUUGUGGUCACUUUUUCCACAUUGUUUUCCCACUGCCAAU .......----.................((((..((((....)--)))((((((..(((((((((...((((((((......)).))))))....)))).)))))...)))))).)))). ( -33.50) >DroYak_CAF1 79024 120 + 1 AUACACAUACAUAUAUUUAACGUAUAUUUUUGAGCUUCUGGUAAAAAGGGUGGGCUGCGGUUGGACCCGUGGCCCAUGUUGUUGUGGUCACUUUUUCCACAUUGUUUUCCUACUGCCAAU .....((...((((((.....))))))...))......(((((...(((((((((..(((......)))..)))))).....(((((.........))))).......)))..))))).. ( -34.40) >consensus AUACACAUACAUAUAUUUAACGUAUAUUUUUGAGCUUCUGAA_AAAAGGGUGGGCUGCGGUUGGACCCGUGGCCCAUGUUGUUGUGGUCACUUUUUCCACAUUGUUUUCCUACUGCCAAU .....................(((.......((((...........((.((((((..(((......)))..)))))).))..(((((.........)))))..))))......))).... (-27.61 = -27.39 + -0.22)

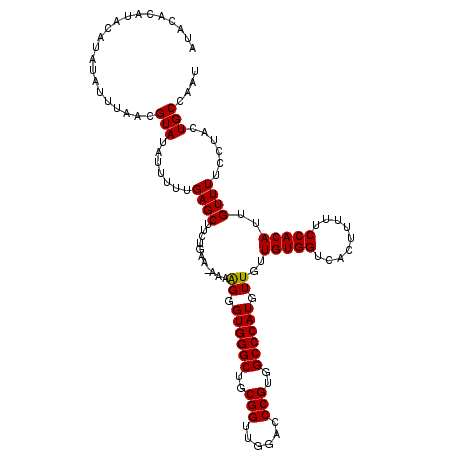
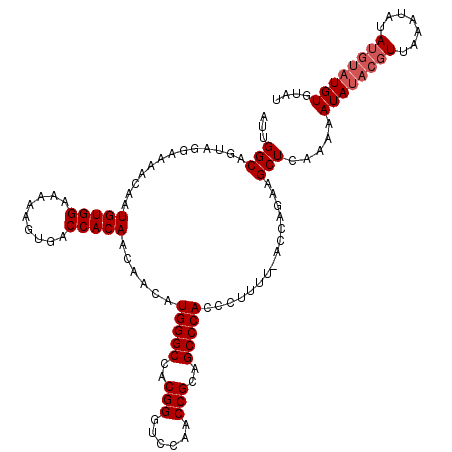
| Location | 16,028,245 – 16,028,360 |
|---|---|
| Length | 115 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 86.94 |
| Mean single sequence MFE | -31.80 |
| Consensus MFE | -22.87 |
| Energy contribution | -24.20 |
| Covariance contribution | 1.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.62 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.61 |
| SVM RNA-class probability | 0.799120 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16028245 115 - 22224390 AUUGGCAAAAGGAAAACAAUGUGGAAAAAGUGACCACAACAACAUGGGCCACGGGUCCAACCGCAGCCCACCCCUUU-UUCGCAAGCUCAAAAAUAUACGUUAAAUAUAUGUAUGU---- .((((.((((((.......(((((.........)))))......(((((..(((......)))..)))))..)))))-).).)))........((((((((.......))))))))---- ( -30.50) >DroEre_CAF1 96466 114 - 1 AUUGGCAGUGGGAAAACAAUGUGGAAAAAGUGACCACAACAACAUGGGCCACGGGUCCAACCGCAGCCCACCCUU--CGGCAGAAGCUCCAAAAUAUACGUUGAAUCUA----UGUGUAU .((((.((((((.......(((((.........)))))......(((((..(((......)))..))))))))((--(....)))))))))).((((((((.......)----))))))) ( -34.70) >DroYak_CAF1 79024 120 - 1 AUUGGCAGUAGGAAAACAAUGUGGAAAAAGUGACCACAACAACAUGGGCCACGGGUCCAACCGCAGCCCACCCUUUUUACCAGAAGCUCAAAAAUAUACGUUAAAUAUAUGUAUGUGUAU .((((....(((.......(((((.........)))))......(((((..(((......)))..))))).))).....))))..........((((((((.......)))))))).... ( -30.20) >consensus AUUGGCAGUAGGAAAACAAUGUGGAAAAAGUGACCACAACAACAUGGGCCACGGGUCCAACCGCAGCCCACCCUUUU_ACCAGAAGCUCAAAAAUAUACGUUAAAUAUAUGUAUGUGUAU ...(((.............(((((.........)))))......(((((..(((......)))..)))))...............))).....((((((((.......)))))))).... (-22.87 = -24.20 + 1.33)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:12:21 2006