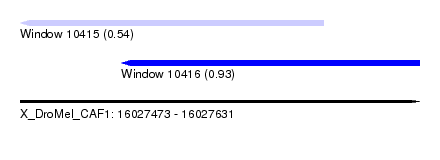
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 16,027,473 – 16,027,631 |
| Length | 158 |
| Max. P | 0.934681 |

| Location | 16,027,473 – 16,027,593 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 86.83 |
| Mean single sequence MFE | -25.90 |
| Consensus MFE | -17.80 |
| Energy contribution | -17.84 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.77 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.01 |
| SVM RNA-class probability | 0.540654 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16027473 120 - 22224390 UGCUGUGAUAACUCAUUCUGCUCAUUUCGCAGGUUGAGCAAAGUCGCAGUUUCAUUUCACAUUAAUGACAAAAGGUUCCCAAAAAAAUAAGAGGGUGAAAUAAAGUUGUCAAAAAGUUAA .((((((((..((((..((((.......))))..))))....))))))))...............((((((...((((((............)))...)))....))))))......... ( -28.50) >DroSec_CAF1 95050 117 - 1 UGCCGUGAUAACUCAUUCUGCUCAUUUCGCAGGUUGAGCAAAGUCGCAGUUUCAUUUCAUAUUAAUGACAAAAGAUCCC--AAAAAA-AAGAGACUGAAAUAAAGUUGUCAAAAAGUUAA .....((((((((((..((((.......))))..))).....((..(((((((...((((....)))).....(....)--......-..)))))))..))...)))))))......... ( -23.30) >DroSim_CAF1 89260 119 - 1 UGCCGUGAUAACUCAUUCUGCUCAUUUCGCAGGUUGAGCAAAGUCGCAGUUUCAUUUCAUAUUAAUGACAAAAGAUCCCCAAAAAAA-AAUAGACUGAAAUAAAGUUGUCAAAAAGUUAA ....(((((..((((..((((.......))))..))))....)))))..................((((....(.....).......-....((((.......))))))))......... ( -19.40) >DroEre_CAF1 95777 116 - 1 UGCCGUGAUAACUCAUUCUGCUCAUUUCGCAGGCUGGGCACAGUCGCAGUUUCAUUUCAUAUUAAUGACAA-AAAGCUGCAAAGAAA--AAA-GGGCAAGUAAAGUUGUCAAAAUGUUAC .....((((((((.....(((((.((((...(((((....)))))(((((((....((((....))))...-.)))))))...))))--...-))))).....))))))))......... ( -33.20) >DroYak_CAF1 78307 112 - 1 UGCCGUGAUAACUCAUUCUGCUCAUUUCGCAGGUUGAGCAAAGUCGCAGUUUCAUCUCAUAUUAAUGACACAAGAUCUACA-----A--AAA-GGCGAAAUAAUGUUGUCAAAAUGUUAA .((((((((..((((..((((.......))))..))))....))))).((.((((.........)))).))..........-----.--...-)))....(((((((.....))))))). ( -25.10) >consensus UGCCGUGAUAACUCAUUCUGCUCAUUUCGCAGGUUGAGCAAAGUCGCAGUUUCAUUUCAUAUUAAUGACAAAAGAUCCCCAAAAAAA_AAAAGGGUGAAAUAAAGUUGUCAAAAAGUUAA ....(((((..((((..((((.......))))..))))....)))))..................((((((..................................))))))......... (-17.80 = -17.84 + 0.04)

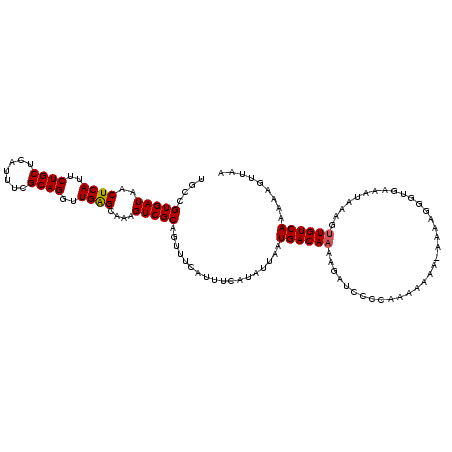

| Location | 16,027,513 – 16,027,631 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.05 |
| Mean single sequence MFE | -30.08 |
| Consensus MFE | -25.32 |
| Energy contribution | -25.00 |
| Covariance contribution | -0.32 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.73 |
| Structure conservation index | 0.84 |
| SVM decision value | 1.24 |
| SVM RNA-class probability | 0.934681 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16027513 118 - 22224390 UUCCGGAUUUUAAUAACAAGUGAGGCCUGACAUCA--CGCUGCUGUGAUAACUCAUUCUGCUCAUUUCGCAGGUUGAGCAAAGUCGCAGUUUCAUUUCACAUUAAUGACAAAAGGUUCCC ....(((..(((((...(((((((((.((....))--.)))((((((((..((((..((((.......))))..))))....)))))))).))))))...)))))...........))). ( -28.72) >DroSec_CAF1 95088 119 - 1 UUCCGGAUUUUAAUAACAAGUGAGGCCUGACAUCACACGCUGCCGUGAUAACUCAUUCUGCUCAUUUCGCAGGUUGAGCAAAGUCGCAGUUUCAUUUCAUAUUAAUGACAAAAGAUCCC- ....(((((((.......((((((((.(((((((((........)))))..((((..((((.......))))..))))....))))..))))))))((((....))))...))))))).- ( -32.30) >DroSim_CAF1 89299 120 - 1 UUCCGGAUUUUAAUAACAAGUGAGGCCUGACAUCACACGCUGCCGUGAUAACUCAUUCUGCUCAUUUCGCAGGUUGAGCAAAGUCGCAGUUUCAUUUCAUAUUAAUGACAAAAGAUCCCC ....(((((((.......((((((((.(((((((((........)))))..((((..((((.......))))..))))....))))..))))))))((((....))))...))))))).. ( -32.30) >DroEre_CAF1 95814 117 - 1 UUCCGGAUUUUAAUAACAAGUGAGGCCUGACAUCA--UGCUGCCGUGAUAACUCAUUCUGCUCAUUUCGCAGGCUGGGCACAGUCGCAGUUUCAUUUCAUAUUAAUGACAA-AAAGCUGC ....((.(((((........))))))).(((((((--((....))))))..((((..((((.......))))..))))....)))(((((((....((((....))))...-.))))))) ( -27.50) >DroYak_CAF1 78339 118 - 1 UUCCGGAUUUUAAUAACAAGUGAGGCCUGACAUCA--UGCUGCCGUGAUAACUCAUUCUGCUCAUUUCGCAGGUUGAGCAAAGUCGCAGUUUCAUCUCAUAUUAAUGACACAAGAUCUAC ....(((((((........(((((((.((((((((--((....))))))..((((..((((.......))))..))))....))))..))))))).((((....))))...))))))).. ( -29.60) >consensus UUCCGGAUUUUAAUAACAAGUGAGGCCUGACAUCA__CGCUGCCGUGAUAACUCAUUCUGCUCAUUUCGCAGGUUGAGCAAAGUCGCAGUUUCAUUUCAUAUUAAUGACAAAAGAUCCCC ....(((((((.......((((((((.((((......(((....)))....((((..((((.......))))..))))....))))..))))))))((((....))))...))))))).. (-25.32 = -25.00 + -0.32)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:12:16 2006