| Sequence ID | X_DroMel_CAF1 |
|---|---|
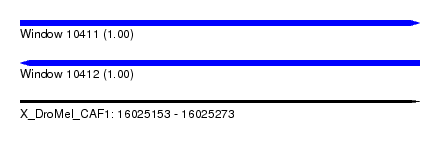
| Location | 16,025,153 – 16,025,273 |
| Length | 120 |
| Max. P | 0.999458 |

| Location | 16,025,153 – 16,025,273 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 80.68 |
| Mean single sequence MFE | -36.08 |
| Consensus MFE | -24.66 |
| Energy contribution | -24.94 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.15 |
| Mean z-score | -4.23 |
| Structure conservation index | 0.68 |
| SVM decision value | 3.59 |
| SVM RNA-class probability | 0.999423 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16025153 120 + 22224390 AUUUUGUCUUCUUAGUAUUACUUCUAGUCCAAAACAUGGACAAAUAAUAGCUCUGCCCGACCAUUGGAACAUUUGAUAAGGUACUUAUGAAAUGUACCAAUGAGCCAGCAGUGCUAUUAU ..........................(((((.....)))))..((((((((.((((..(..((((((.((((((.(((((...))))).)))))).))))))..)..)))).)))))))) ( -40.00) >DroSec_CAF1 92829 116 + 1 AUUUCCUCUUCUUAGGAUUACUUCUAUUCCAAGACAUGGACAAGUAAUAGCUCUGCCCGACCAUUGGAACAUUUCAUAAGGUACUUAUGAAAUGUACCAAUGUGCCAGCAGUGCAA---- ...((((......))))((((((....((((.....)))).))))))..((.((((..(..((((((.((((((((((((...)))))))))))).))))))..)..)))).))..---- ( -40.00) >DroSim_CAF1 85861 115 + 1 AUUUCCUUUUUUUAGAAUUACUUCUAU-CCAAAACAUGGACCAGUAAUAGCUUUGCCCGACCAUUGGAACAUUUCAUAAGGUACUUAUGAAAUGUACAAAUGAGCCAGCAGUGCUG---- ................((((((....(-(((.....))))..))))))(((.((((..(..((((.(.((((((((((((...)))))))))))).).))))..)..)))).))).---- ( -29.20) >DroEre_CAF1 93425 115 + 1 CUUUCUUGGUCUCAGUACUACUCCAUCUCCAGAACAUGAACUAA-AGCAGCUCUGCCAGACCAUUGGACCCUUUGAUAAGGUACUUAUGAAAGGUACCAAUGGGCCCGCAGUGCUA---- .....(((((.(((((.((...........)).)).))))))))-...(((.((((....(((((((..(((((.(((((...))))).)))))..)))))))....)))).))).---- ( -35.40) >DroYak_CAF1 76074 114 + 1 UUAACUUGUUCUCAAUAUUACUCCUAUUCCAAAACAAGGACUAA-GACAGCUCUGCCGGACCAUUGGAACCU-GGAUAAGGCACUUAUGAAAGGUACCAAUGAGCCCGCAGUGCUG---- ....((((((...((((.......))))....))))))......-..((((.((((.((..((((((.((((-..(((((...)))))...)))).))))))..)).)))).))))---- ( -35.80) >consensus AUUUCCUCUUCUUAGUAUUACUUCUAUUCCAAAACAUGGACAAAUAAUAGCUCUGCCCGACCAUUGGAACAUUUGAUAAGGUACUUAUGAAAUGUACCAAUGAGCCAGCAGUGCUA____ ...............................................((((.((((..(..((((((.((((((.(((((...))))).)))))).))))))..)..)))).)))).... (-24.66 = -24.94 + 0.28)

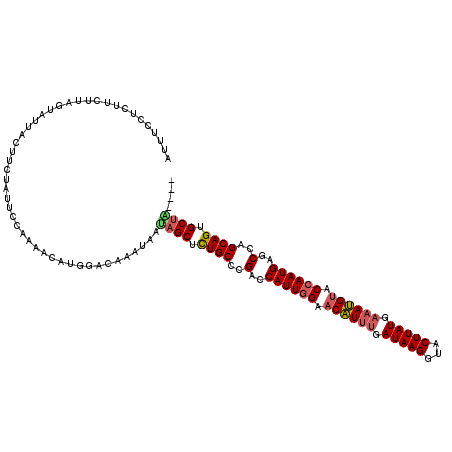

| Location | 16,025,153 – 16,025,273 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 80.68 |
| Mean single sequence MFE | -40.96 |
| Consensus MFE | -31.08 |
| Energy contribution | -31.32 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.18 |
| Mean z-score | -4.45 |
| Structure conservation index | 0.76 |
| SVM decision value | 3.62 |
| SVM RNA-class probability | 0.999458 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16025153 120 - 22224390 AUAAUAGCACUGCUGGCUCAUUGGUACAUUUCAUAAGUACCUUAUCAAAUGUUCCAAUGGUCGGGCAGAGCUAUUAUUUGUCCAUGUUUUGGACUAGAAGUAAUACUAAGAAGACAAAAU ((((((((.(((((.(..((((((.((((((.(((((...))))).)))))).))))))..).))))).))))))))..(((((.....))))).......................... ( -41.90) >DroSec_CAF1 92829 116 - 1 ----UUGCACUGCUGGCACAUUGGUACAUUUCAUAAGUACCUUAUGAAAUGUUCCAAUGGUCGGGCAGAGCUAUUACUUGUCCAUGUCUUGGAAUAGAAGUAAUCCUAAGAAGAGGAAAU ----..((.((((((((.((((((.((((((((((((...)))))))))))).))))))))).))))).))..((((((.((((.....))))....))))))((((......))))... ( -45.90) >DroSim_CAF1 85861 115 - 1 ----CAGCACUGCUGGCUCAUUUGUACAUUUCAUAAGUACCUUAUGAAAUGUUCCAAUGGUCGGGCAAAGCUAUUACUGGUCCAUGUUUUGG-AUAGAAGUAAUUCUAAAAAAAGGAAAU ----.(((..((((.(..((((.(.((((((((((((...)))))))))))).).))))..).))))..))).(((((.(((((.....)))-))...)))))((((......))))... ( -36.80) >DroEre_CAF1 93425 115 - 1 ----UAGCACUGCGGGCCCAUUGGUACCUUUCAUAAGUACCUUAUCAAAGGGUCCAAUGGUCUGGCAGAGCUGCU-UUAGUUCAUGUUCUGGAGAUGGAGUAGUACUGAGACCAAGAAAG ----.....((((.((.(((((((..(((((.(((((...))))).)))))..))))))).)).)))).((((((-(((.((((.....))))..)))))))))................ ( -40.40) >DroYak_CAF1 76074 114 - 1 ----CAGCACUGCGGGCUCAUUGGUACCUUUCAUAAGUGCCUUAUCC-AGGUUCCAAUGGUCCGGCAGAGCUGUC-UUAGUCCUUGUUUUGGAAUAGGAGUAAUAUUGAGAACAAGUUAA ----((((.((((((((.((((((.((((...(((((...)))))..-)))).)))))))))).)))).))))((-(((.(((.......))).)))))..................... ( -39.80) >consensus ____UAGCACUGCUGGCUCAUUGGUACAUUUCAUAAGUACCUUAUCAAAUGUUCCAAUGGUCGGGCAGAGCUAUUAUUAGUCCAUGUUUUGGAAUAGAAGUAAUACUAAGAACAAGAAAU .....(((.((((((((.((((((.((((((.(((((...))))).)))))).)))))))))).)))).)))((((((..((((.....)))).....))))))................ (-31.08 = -31.32 + 0.24)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:12:12 2006