
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 16,021,389 – 16,021,720 |
| Length | 331 |
| Max. P | 0.971034 |

| Location | 16,021,389 – 16,021,508 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 98.66 |
| Mean single sequence MFE | -28.84 |
| Consensus MFE | -26.22 |
| Energy contribution | -26.62 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.65 |
| Structure conservation index | 0.91 |
| SVM decision value | 1.02 |
| SVM RNA-class probability | 0.901025 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16021389 119 + 22224390 AAAUGAGUUCAAUUUGUUAGUGCUGCAGACACACACGUACUAAACACACUAAUUUGCGGCAUCGCUUCUGGACAUAUCAGCAAUUUGUUGCCAGCAGCUGUCCUCAGACGAACUU-UUAC ....((((((.........((((((((((.......((.....)).......)))))))))).(((.((((.((...(((....))).)))))).))).(((....)))))))))-.... ( -29.04) >DroSec_CAF1 89092 119 + 1 AAAUGAGUUCAAUUUGUUAGUGCUGCAGACACACACGUACUAAACACACUAAUUUGCGGCAUCGCUUCUGGACAUAUCAGCAAUUUGUUGCCAGCAGCUGUCCUCAGACGAACUU-UUAC ....((((((.........((((((((((.......((.....)).......)))))))))).(((.((((.((...(((....))).)))))).))).(((....)))))))))-.... ( -29.04) >DroSim_CAF1 83136 119 + 1 AAAUGAGUUCAAUUUGUUAGUGCUGCAGACACACACGUACUAAACACACUAAUUUGCGGCAUCGCUUCUGGACAUAUCAGCAAUUUGUUGCCAGCAGCUGUCCUCAGACGAACUU-UUAC ....((((((.........((((((((((.......((.....)).......)))))))))).(((.((((.((...(((....))).)))))).))).(((....)))))))))-.... ( -29.04) >DroEre_CAF1 87138 119 + 1 AAAUGAGUUCAAUUUGUUAGUGCUGCAGACACACACGUACUAAACACACUAAUUUGCGGCAUCGCUUCUGGACAUAUCAGCAAUUUGUUUCCAGCAGCUGUCCUCAGCCGAACUU-UUAC ....((((((.........((((((((((.......((.....)).......)))))))))).(((.(((((.(((.........))).))))).)))...........))))))-.... ( -28.54) >DroYak_CAF1 72688 120 + 1 AAAUGAGUUCAAUUUGUUAGUGCUGCAGACACACACGUACUAAACACACUAAUUUGCGGCAUCGCUUCUGGACAUAUCAGCAAUUUGUUUCCAGCAGCUGUCCUCAGCCGAACUUUUUAC ....((((((.........((((((((((.......((.....)).......)))))))))).(((.(((((.(((.........))).))))).)))...........))))))..... ( -28.54) >consensus AAAUGAGUUCAAUUUGUUAGUGCUGCAGACACACACGUACUAAACACACUAAUUUGCGGCAUCGCUUCUGGACAUAUCAGCAAUUUGUUGCCAGCAGCUGUCCUCAGACGAACUU_UUAC ....((((((.........((((((((((.......((.....)).......)))))))))).(((.((((......((((.....)))))))).)))...........))))))..... (-26.22 = -26.62 + 0.40)

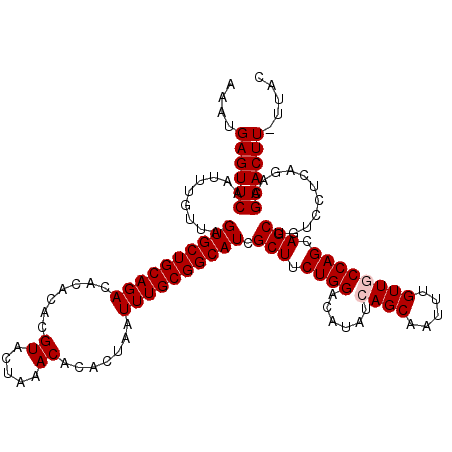
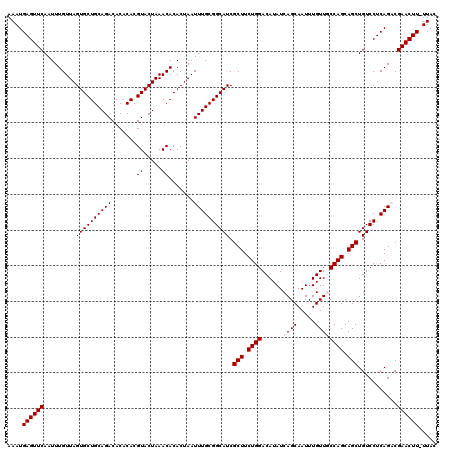
| Location | 16,021,429 – 16,021,536 |
|---|---|
| Length | 107 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.23 |
| Mean single sequence MFE | -26.07 |
| Consensus MFE | -22.37 |
| Energy contribution | -22.61 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.69 |
| Structure conservation index | 0.86 |
| SVM decision value | 1.36 |
| SVM RNA-class probability | 0.946053 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16021429 107 + 22224390 UAAACACACUAAUUUGCGGCAUCGCUUCUGGACAUAUCAGCAAUUUGUUGCCAGCAGCUGUCCUCAGACGAACUU-UUACAGGACUUUCGUAAAUUUCUGGUCCUCUU------------ ......((..(((((((((....(((.((((.((...(((....))).)))))).))).(((((...........-....)))))..)))))))))..))........------------ ( -23.86) >DroSec_CAF1 89132 107 + 1 UAAACACACUAAUUUGCGGCAUCGCUUCUGGACAUAUCAGCAAUUUGUUGCCAGCAGCUGUCCUCAGACGAACUU-UUACAGGACUUUCGUAAAUUUCUGGUCCUGUU------------ ............((((.((.((.(((.((((.((...(((....))).)))))).))).)))).)))).......-..((((((((.............)))))))).------------ ( -25.62) >DroSim_CAF1 83176 107 + 1 UAAACACACUAAUUUGCGGCAUCGCUUCUGGACAUAUCAGCAAUUUGUUGCCAGCAGCUGUCCUCAGACGAACUU-UUACAGGACUUUCGUAAAUUUCUGGUCCUGUU------------ ............((((.((.((.(((.((((.((...(((....))).)))))).))).)))).)))).......-..((((((((.............)))))))).------------ ( -25.62) >DroEre_CAF1 87178 107 + 1 UAAACACACUAAUUUGCGGCAUCGCUUCUGGACAUAUCAGCAAUUUGUUUCCAGCAGCUGUCCUCAGCCGAACUU-UUACAGGACUUUCGUAAGUUUCUGGUCCUGUU------------ ................((((...(((.(((((.(((.........))).))))).)))((....)))))).....-..((((((((..(....).....)))))))).------------ ( -29.10) >DroYak_CAF1 72728 120 + 1 UAAACACACUAAUUUGCGGCAUCGCUUCUGGACAUAUCAGCAAUUUGUUUCCAGCAGCUGUCCUCAGCCGAACUUUUUACAGGACUUUCGUAAAUUUCUGUUCCUGUUCCUGUUCCUGUU ..((((...........(((...(((.(((((.(((.........))).))))).)))((....)))))((((.....(((((((..............).))))))....)))).)))) ( -26.14) >consensus UAAACACACUAAUUUGCGGCAUCGCUUCUGGACAUAUCAGCAAUUUGUUGCCAGCAGCUGUCCUCAGACGAACUU_UUACAGGACUUUCGUAAAUUUCUGGUCCUGUU____________ ......((..(((((((((....(((.((((......((((.....)))))))).))).(((((................)))))..)))))))))..)).................... (-22.37 = -22.61 + 0.24)

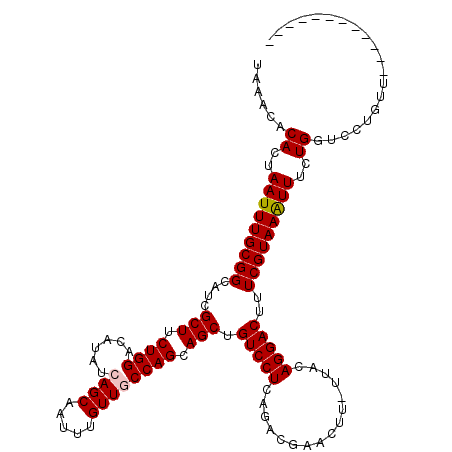
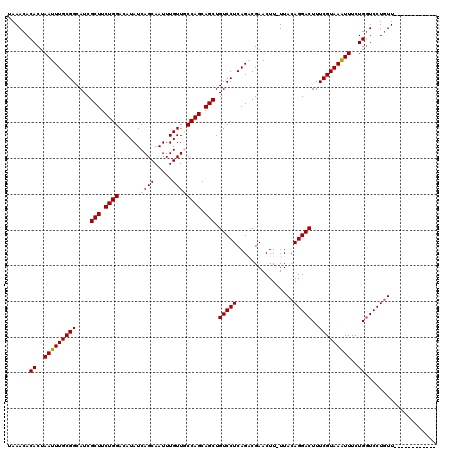
| Location | 16,021,536 – 16,021,650 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.07 |
| Mean single sequence MFE | -23.05 |
| Consensus MFE | -20.18 |
| Energy contribution | -20.22 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.73 |
| Structure conservation index | 0.88 |
| SVM decision value | 1.67 |
| SVM RNA-class probability | 0.971034 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16021536 114 + 22224390 ------CCGGUCAGUCCAGCUCAAACUUGAUCCGUCGAGCGUUGACCAUAUUUCUUCAACUCGGCACCCGCCAAGCAAAUCGACAAAACCAAUAAAACAAAACAAUUUUGCGGCCAAAAA ------..((((.(((.((......)).)))..((((((((((((..........)))))..(((....)))..))...)))))...........................))))..... ( -22.90) >DroSec_CAF1 89239 114 + 1 ------CCGGUCAGUUCAGCUCAAACUUGAUCCGUCGAGCGUUGACCAUAUUUCUUCAACUCGGCACCCGCCAAGCAAAUCGACAAAACCAAUAAAACAAAACAAUUUUGCGGCCAAAAA ------..((((...((((.......))))...((((((((((((..........)))))..(((....)))..))...)))))...........................))))..... ( -22.20) >DroSim_CAF1 83283 114 + 1 ------CCGGUCAGUUCAGCUCAAACUUGAUCCGUUGAGCGUUGACCAUAUUUCUUCAACUCGGCACCCGCCAAGCAAAUCGACAAAACCAAUAAAACAAAACAAUUUUGCGGCCAAAAA ------..((((.((((((((((....)))...)))))))(((((..........)))))..(((....)))..(((((...........................)))))))))..... ( -24.13) >DroEre_CAF1 87285 114 + 1 ------CCGGUCAGUCCUGCUCAGACUUGAUCCGUCGAGCGUUGACCAUAUUUCCUCAACUCGGCACCCGCCAAGCAAAUCGACAAAACCAAUAAAACAAAACAAUUUUGCGGCCAAAAG ------..((((.(((..((((.(((.......)))))))(((((..........)))))..(((....))).........))).............((((.....)))).))))..... ( -24.50) >DroYak_CAF1 72848 120 + 1 CCUGUUUCGGUCAGUCCAACUCAAAGUUGAUCCGUCGAGCGUUGACCAUAUUUCAUCAACUCGACACCCGCCAAGCAAAUCGACAAAACCAAUAAAACAAAACAAUUUUGCGGCCAAAAA ........((((.(((((((.....))))....((((((...(((.......)))....))))))................))).............((((.....)))).))))..... ( -21.50) >consensus ______CCGGUCAGUCCAGCUCAAACUUGAUCCGUCGAGCGUUGACCAUAUUUCUUCAACUCGGCACCCGCCAAGCAAAUCGACAAAACCAAUAAAACAAAACAAUUUUGCGGCCAAAAA ........((((.....................((((((((((((..........)))))..(((....)))..))...))))).............((((.....)))).))))..... (-20.18 = -20.22 + 0.04)

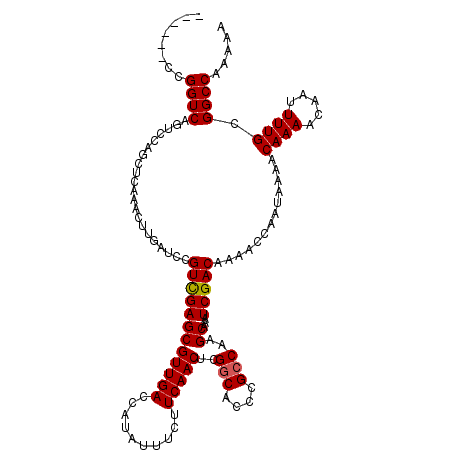
| Location | 16,021,570 – 16,021,686 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.12 |
| Mean single sequence MFE | -35.08 |
| Consensus MFE | -29.12 |
| Energy contribution | -28.36 |
| Covariance contribution | -0.76 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.42 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.26 |
| SVM RNA-class probability | 0.659533 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16021570 116 - 22224390 CAUACCUAU----UUCCCGGGCCAGGGAAGCUCUCUUCGCUUUUUGGCCGCAAAAUUGUUUUGUUUUAUUGGUUUUGUCGAUUUGCUUGGCGGGUGCCGAGUUGAAGAAAUAUGGUCAAC ...((((((----(((.(((((((((.((((.......)))))))))))(((((((((...........))))))))))).((.(((((((....))))))).)).)))))).))).... ( -34.50) >DroSec_CAF1 89273 115 - 1 CACACCUAU----UUCCCGGGCCAGGGAAGCUCUC-UCGCUUUUUGGCCGCAAAAUUGUUUUGUUUUAUUGGUUUUGUCGAUUUGCUUGGCGGGUGCCGAGUUGAAGAAAUAUGGUCAAC ...((((((----(((.(((((((((.((((....-..)))))))))))(((((((((...........))))))))))).((.(((((((....))))))).)).)))))).))).... ( -35.10) >DroSim_CAF1 83317 115 - 1 CACACCUAU----UUCCCGGGCCAGGGAAGCUCUC-UCGCUUUUUGGCCGCAAAAUUGUUUUGUUUUAUUGGUUUUGUCGAUUUGCUUGGCGGGUGCCGAGUUGAAGAAAUAUGGUCAAC ...((((((----(((.(((((((((.((((....-..)))))))))))(((((((((...........))))))))))).((.(((((((....))))))).)).)))))).))).... ( -35.10) >DroEre_CAF1 87319 116 - 1 CACACCUAU---UUUCCCGGGCCAGAAGAGCUCUC-UCGCCUUUUGGCCGCAAAAUUGUUUUGUUUUAUUGGUUUUGUCGAUUUGCUUGGCGGGUGCCGAGUUGAGGAAAUAUGGUCAAC ...(((...---((((((((((((((((.((....-..)))))))))))(((((((((...........))))))))))).((.(((((((....))))))).)))))))...))).... ( -39.20) >DroYak_CAF1 72888 119 - 1 CACACCUAUGUAUUUUCCGGGGCAGAAAAGCUCAC-UCGCUUUUUGGCCGCAAAAUUGUUUUGUUUUAUUGGUUUUGUCGAUUUGCUUGGCGGGUGUCGAGUUGAUGAAAUAUGGUCAAC .((((((...........(.(((.(((((((....-..))))))).))).)......(((..((...(((((.....)))))..))..)))))))))...((((((.(....).)))))) ( -31.50) >consensus CACACCUAU____UUCCCGGGCCAGGGAAGCUCUC_UCGCUUUUUGGCCGCAAAAUUGUUUUGUUUUAUUGGUUUUGUCGAUUUGCUUGGCGGGUGCCGAGUUGAAGAAAUAUGGUCAAC ...(((...........(((((((((((.((.......)))))))))))(((((((((...........))))))))))).((.(((((((....))))))).))........))).... (-29.12 = -28.36 + -0.76)

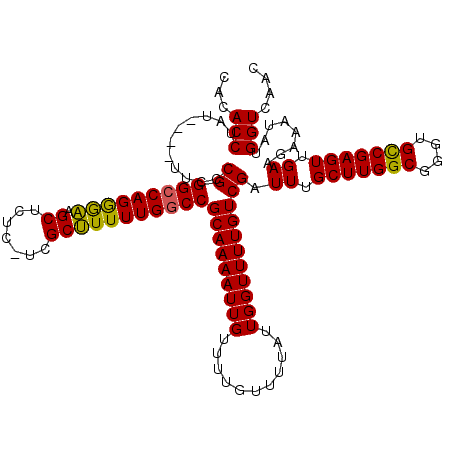

| Location | 16,021,610 – 16,021,720 |
|---|---|
| Length | 110 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.79 |
| Mean single sequence MFE | -21.99 |
| Consensus MFE | -19.76 |
| Energy contribution | -19.16 |
| Covariance contribution | -0.60 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.46 |
| Structure conservation index | 0.90 |
| SVM decision value | 1.11 |
| SVM RNA-class probability | 0.916530 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16021610 110 - 22224390 UCAUUUAAUAACCUCAACAACC------CCUCAAUAAAAACAUACCUAU----UUCCCGGGCCAGGGAAGCUCUCUUCGCUUUUUGGCCGCAAAAUUGUUUUGUUUUAUUGGUUUUGUCG ................((((..------((..(((((((....((..((----((..(((.(((((.((((.......))))))))))))..)))).))....)))))))))..)))).. ( -21.40) >DroSec_CAF1 89313 109 - 1 UCAUUUAAUAACCUCAACAACC------CCUCAAUAAAAACACACCUAU----UUCCCGGGCCAGGGAAGCUCUC-UCGCUUUUUGGCCGCAAAAUUGUUUUGUUUUAUUGGUUUUGUCG ................((((..------((..((((((...........----......(((((((.((((....-..)))))))))))(((((.....)))))))))))))..)))).. ( -21.30) >DroSim_CAF1 83357 109 - 1 UCAUUUAAUAACCUCAACAACC------CCUCAAUAAAAACACACCUAU----UUCCCGGGCCAGGGAAGCUCUC-UCGCUUUUUGGCCGCAAAAUUGUUUUGUUUUAUUGGUUUUGUCG ................((((..------((..((((((...........----......(((((((.((((....-..)))))))))))(((((.....)))))))))))))..)))).. ( -21.30) >DroEre_CAF1 87359 116 - 1 UCAUUUAAUAACCUCAACAACCCCAGCCCCUCAAUAAAAACACACCUAU---UUUCCCGGGCCAGAAGAGCUCUC-UCGCCUUUUGGCCGCAAAAUUGUUUUGUUUUAUUGGUUUUGUCG ................((((..((((.......................---.......(((((((((.((....-..)))))))))))(((((.....)))))....))))..)))).. ( -24.80) >DroYak_CAF1 72928 113 - 1 UCAUUUAAUAACCUCAACAACC------CCUCAAUAAAAACACACCUAUGUAUUUUCCGGGGCAGAAAAGCUCAC-UCGCUUUUUGGCCGCAAAAUUGUUUUGUUUUAUUGGUUUUGUCG ................((((..------((......((((((((....))).......(.(((.(((((((....-..))))))).))).)......)))))........))..)))).. ( -21.14) >consensus UCAUUUAAUAACCUCAACAACC______CCUCAAUAAAAACACACCUAU____UUCCCGGGCCAGGGAAGCUCUC_UCGCUUUUUGGCCGCAAAAUUGUUUUGUUUUAUUGGUUUUGUCG ..............................((((((((.....................(((((((((.((.......)))))))))))(((((.....)))))))))))))........ (-19.76 = -19.16 + -0.60)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:12:08 2006