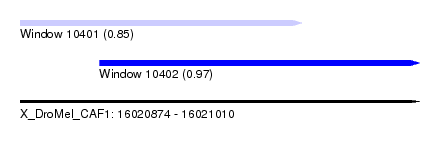
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 16,020,874 – 16,021,010 |
| Length | 136 |
| Max. P | 0.968031 |

| Location | 16,020,874 – 16,020,970 |
|---|---|
| Length | 96 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 88.83 |
| Mean single sequence MFE | -35.64 |
| Consensus MFE | -31.02 |
| Energy contribution | -30.98 |
| Covariance contribution | -0.04 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.57 |
| Structure conservation index | 0.87 |
| SVM decision value | 0.78 |
| SVM RNA-class probability | 0.848904 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
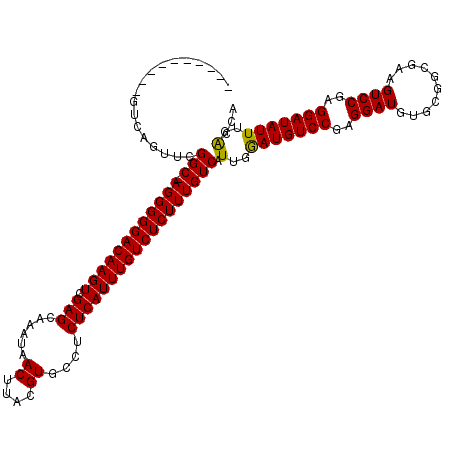
>X_DroMel_CAF1 16020874 96 + 22224390 UGACGGCACGGGGAUGGUGGCAUGAUG-----------------------GUCAGUUCGGC-AGGGGGACAAGUCGAGCAAAUAACUUACGUGCCUCUCAUUUGUCUCUUUGUCAUUGAA ....((((.((((((((.((((((..(-----------------------((..(((((((-..(....)..))))))).....)))..)))))).)).....)))))).))))...... ( -33.90) >DroSec_CAF1 88593 96 + 1 UGACGGCACGGGGAUGGUGGCACGAUG-----------------------GUCAGUUCGGC-AGGGGGACAAGUCGAGCAAAUAACUUACGUGCCUCUCAUUUGUCUCUUUGUCAUUGGA ....((((.((((((((.((((((..(-----------------------((..(((((((-..(....)..))))))).....)))..)))))).)).....)))))).))))...... ( -35.70) >DroSim_CAF1 82625 96 + 1 UGACGGCACGGGGAUGGUGGCACGAUG-----------------------GUCAGUUCGGC-AGGGGGACAAGUCGAGCAAAUAACUUACGUGCCUCUCAUUUGUCUCUUUGUCAUUGGA ....((((.((((((((.((((((..(-----------------------((..(((((((-..(....)..))))))).....)))..)))))).)).....)))))).))))...... ( -35.70) >DroEre_CAF1 86614 96 + 1 UGACGGCACGGGGAUGGUGGCAUGAUG-----------------------GUAAGUGCGGC-AGGGGGACAAGUCGAGCAAAUAACUUACGUGCCUCUCAUUUGUCUCUUUGUCAUUGGA ....((((.((((((((.((((((..(-----------------------((...((((((-..(....)..)))..)))....)))..)))))).)).....)))))).))))...... ( -30.10) >DroYak_CAF1 72093 120 + 1 UGACGGCACGGGGAUGGUGGCAUGGUGGCAUGGUGGCACGAUGGUCAGUGGUCAGUUCGGCGAGGGGGACAAGUCGAGCAAAUAACUUACGUGCCUCUCAUUUGUCUCUUUGUCAUUGGA ....((((.(((((..((((((((....))))(.((((((.((..(...)..))(((((((...(....)..)))))))..........)))))).)))).)..))))).))))...... ( -42.80) >consensus UGACGGCACGGGGAUGGUGGCAUGAUG_______________________GUCAGUUCGGC_AGGGGGACAAGUCGAGCAAAUAACUUACGUGCCUCUCAUUUGUCUCUUUGUCAUUGGA ....((((.((((((((.((((((..............................(((((((...(....)..)))))))((.....)).)))))).)).....)))))).))))...... (-31.02 = -30.98 + -0.04)


| Location | 16,020,901 – 16,021,010 |
|---|---|
| Length | 109 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.36 |
| Mean single sequence MFE | -38.58 |
| Consensus MFE | -34.44 |
| Energy contribution | -34.12 |
| Covariance contribution | -0.32 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.64 |
| Structure conservation index | 0.89 |
| SVM decision value | 1.62 |
| SVM RNA-class probability | 0.968031 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16020901 109 + 22224390 ----------GUCAGUUCGGC-AGGGGGACAAGUCGAGCAAAUAACUUACGUGCCUCUCAUUUGUCUCUUUGUCAUUGAAUGUCCGAGGAUGUGCGGCGAAGUCCGAGGAUAUUUGACCA ----------(((.....(((-((((((((((((.(((......((....))....))))))))))))))))))...((((((((..((((..........))))..))))))))))).. ( -38.60) >DroSec_CAF1 88620 109 + 1 ----------GUCAGUUCGGC-AGGGGGACAAGUCGAGCAAAUAACUUACGUGCCUCUCAUUUGUCUCUUUGUCAUUGGAUGUCCGAGGAUGUGCGGCGAAGUCCGAGGAUAUUUGACCA ----------((((....(((-((((((((((((.(((......((....))....))))))))))))))))))....(((((((..((((..........))))..))))))))))).. ( -38.20) >DroSim_CAF1 82652 109 + 1 ----------GUCAGUUCGGC-AGGGGGACAAGUCGAGCAAAUAACUUACGUGCCUCUCAUUUGUCUCUUUGUCAUUGGAUGUCCGAGGAUGUGCGGCGAAGUCCGAGGAUAUUUGACCA ----------((((....(((-((((((((((((.(((......((....))....))))))))))))))))))....(((((((..((((..........))))..))))))))))).. ( -38.20) >DroEre_CAF1 86641 109 + 1 ----------GUAAGUGCGGC-AGGGGGACAAGUCGAGCAAAUAACUUACGUGCCUCUCAUUUGUCUCUUUGUCAUUGGAUGUCCGAGGAUGUGCGGCGAAGUCCGAGGAUAUUUGGCCG ----------........(((-((((((((((((.(((......((....))....)))))))))))))))))).(..(((((((..((((..........))))..)))))))..)... ( -38.00) >DroYak_CAF1 72133 120 + 1 AUGGUCAGUGGUCAGUUCGGCGAGGGGGACAAGUCGAGCAAAUAACUUACGUGCCUCUCAUUUGUCUCUUUGUCAUUGGAUGUCCGAGGAUGUGCGGCGAAGUCCGAGGAUAUUUGACCA .(((((((..(((...(((((((((((.((((((..........))))..)).)))))).((((((.(..((((.((((....)))).)))).).))))))).)))).)))..))))))) ( -39.90) >consensus __________GUCAGUUCGGC_AGGGGGACAAGUCGAGCAAAUAACUUACGUGCCUCUCAUUUGUCUCUUUGUCAUUGGAUGUCCGAGGAUGUGCGGCGAAGUCCGAGGAUAUUUGACCA ..................(((.((((((((((((.(((......((....))....)))))))))))))))))).(..(((((((..((((..........))))..)))))))..)... (-34.44 = -34.12 + -0.32)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:12:03 2006