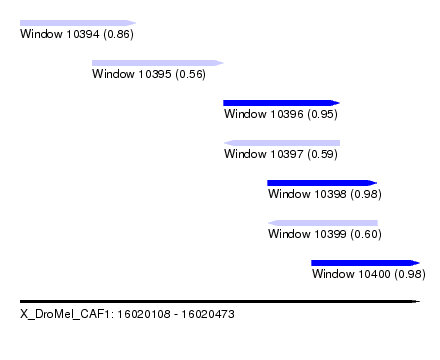
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 16,020,108 – 16,020,473 |
| Length | 365 |
| Max. P | 0.983720 |

| Location | 16,020,108 – 16,020,214 |
|---|---|
| Length | 106 |
| Sequences | 4 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 91.63 |
| Mean single sequence MFE | -37.58 |
| Consensus MFE | -31.91 |
| Energy contribution | -32.10 |
| Covariance contribution | 0.19 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.88 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.84 |
| SVM RNA-class probability | 0.862854 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16020108 106 + 22224390 GGUUUGGGUUUUCAGUUUUGGGUUUUUGGAUU------UGGGGAAUGGGCUUCAGUU-CGAGCUGAGGUCUCGUGUCCGAAAAGCGGAGCGGUAGAGCGGAUCGUUCUCCGGC .((((.(.(((((....(..((((....))))------..)(((((((((((((((.-...))))))))).))).)))))))).).))))((.((((((...))))))))... ( -38.00) >DroSec_CAF1 87860 106 + 1 GGUUUGGGUUUUCAGUUUUGGGUUUUUGGAUU------UGGGGAAUGGGCUUCAGUG-CGGGCUGAGGUCUCGUGUCCGAAAAGCGGAGCGGUAGAGCGGAUCGAUCUCCGGC .((((.(.(((((....(..((((....))))------..)(((((((((((((((.-...))))))))).))).)))))))).).)))).......((((......)))).. ( -34.00) >DroSim_CAF1 81913 106 + 1 GGUUUGGGUUUUCAGUUUUGGAUUUUUGGAUU------UGGGGAAUGGGCUUCAGUG-CGAGCUGAGGUCUCGUGUCCGAAAAGCGGAGCGGUAGAGCGGAUCGUUCUGCGGC .((((.(.(((((....(..((((....))))------..)(((((((((((((((.-...))))))))).))).)))))))).).)))).((((((((...))))))))... ( -40.00) >DroEre_CAF1 85866 113 + 1 GGUUUGGGUUUUCAGUUUUGGGUUUUAGGUUUUGGGUCUUGGGAAUGGGCUUUAGCUUGGAGCUGAGGUCUCGUGUCCGAAAAGCGGAGCGGUAGAGCGGAUCGUUCUCCGGC .....(((.(((((((((..((((..((((((....((....))..)))))).))))..))))))))).)))...((((.....)))).(((.((((((...))))))))).. ( -38.30) >consensus GGUUUGGGUUUUCAGUUUUGGGUUUUUGGAUU______UGGGGAAUGGGCUUCAGUG_CGAGCUGAGGUCUCGUGUCCGAAAAGCGGAGCGGUAGAGCGGAUCGUUCUCCGGC .((((.(.(((((.......((((....))))........((..((((((((((((.....))))))))).)))..))))))).).))))((.((((((...))))))))... (-31.91 = -32.10 + 0.19)

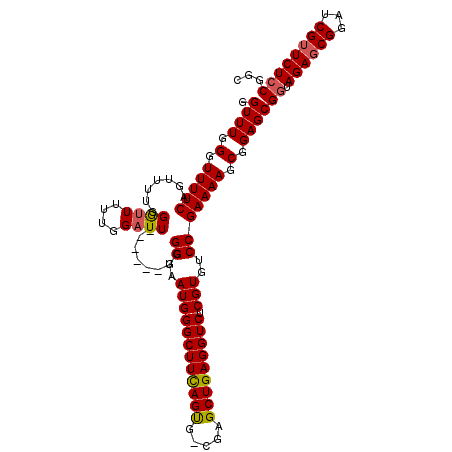
| Location | 16,020,174 – 16,020,294 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 98.33 |
| Mean single sequence MFE | -35.82 |
| Consensus MFE | -32.90 |
| Energy contribution | -33.10 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.27 |
| Structure conservation index | 0.92 |
| SVM decision value | 0.06 |
| SVM RNA-class probability | 0.562933 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16020174 120 + 22224390 UGUCCGAAAAGCGGAGCGGUAGAGCGGAUCGUUCUCCGGCGUGGAGGGAAAGCGUGAAGCGUAUAAAGUCAUAAGGGUUUAUGCAUGAAUUGAUUGCAUCAGCGUGUGCUGGUGGUAGUU ..((((.....))))((((.((((((...)))))))).((((.........))))...))(((((((.((....)).))))))).......((((((((((((....)))))).)))))) ( -35.80) >DroSec_CAF1 87926 120 + 1 UGUCCGAAAAGCGGAGCGGUAGAGCGGAUCGAUCUCCGGCGUGGAGGGAAAGCGUGAAGCGUAUAAAGUCAUAAGGGUUUAUGCAUGAAUUGAUUGCAUCAGCGUGUGCUGGUGGUAGUU ..((((.....))))((......)).(((((((((((.....)))))....((.....))(((((((.((....)).))))))).....)))))).(((((((....)))))))...... ( -34.30) >DroSim_CAF1 81979 120 + 1 UGUCCGAAAAGCGGAGCGGUAGAGCGGAUCGUUCUGCGGCGUGGAGGGAAAGCGUGAAGCGUAUAAAGUCAUAAGGGUUUAUGCAUGAAUUGAUUGCAUCAGCGUGUGCUGGUGGUAGUU (.((((....((((((((((.......))))))))))....)))).)....((.....))(((((((.((....)).))))))).......((((((((((((....)))))).)))))) ( -39.60) >DroEre_CAF1 85939 118 + 1 UGUCCGAAAAGCGGAGCGGUAGAGCGGAUCGUUCUCCGGC--GGAGGGAAAGCGUGAAGCGUAUAAAGUCAUAAGGGUUUAUGCAUGAAUUGAUUGCAUCAGCGUGUGCUGGUGGUAGUU (.((((....(.((((((((.......)))))))).)..)--))).)....((.....))(((((((.((....)).))))))).......((((((((((((....)))))).)))))) ( -34.70) >DroYak_CAF1 71400 118 + 1 UGUCCGAAAAGCGGAGCGGUAGAGCGGAUCGUUCUCCGGC--GGAGGGAAAGCGUGAAGCGUAUAAAGUCAUAAGGGUUUAUGCAUGAAUUGAUUGCAUCAGCGUGUGCUGGUGGUAGUU (.((((....(.((((((((.......)))))))).)..)--))).)....((.....))(((((((.((....)).))))))).......((((((((((((....)))))).)))))) ( -34.70) >consensus UGUCCGAAAAGCGGAGCGGUAGAGCGGAUCGUUCUCCGGCGUGGAGGGAAAGCGUGAAGCGUAUAAAGUCAUAAGGGUUUAUGCAUGAAUUGAUUGCAUCAGCGUGUGCUGGUGGUAGUU ..((((.....))))((.(.((((((...)))))).).))...........((.....))(((((((.((....)).))))))).......((((((((((((....)))))).)))))) (-32.90 = -33.10 + 0.20)



| Location | 16,020,294 – 16,020,400 |
|---|---|
| Length | 106 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.31 |
| Mean single sequence MFE | -33.52 |
| Consensus MFE | -26.30 |
| Energy contribution | -26.30 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.24 |
| Structure conservation index | 0.78 |
| SVM decision value | 1.37 |
| SVM RNA-class probability | 0.947037 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16020294 106 + 22224390 GUACUAACGAAAUAAGAAUUAUAUGCCUUGUGUAGCAAUUCGCUUUAAUAAUCUAUGAAGUGGAUGGGCAGUGGCUUCUUGCCACUUUCGCCUCAAGUGUGG--------------CAAG ........((((...........(((((((.....))((((((((((........)))))))))))))))(((((.....)))))))))(((.(....).))--------------)... ( -30.00) >DroSec_CAF1 88046 106 + 1 GUAAUAACGAAAUAAGAAUUAUAUGCCUUGUGUAGCAAUUCGCUUUAAUAAUCUAUGAAGUGGAUGGGCAGUGGCUUCUUGCCACUUUCGCCUCAAGUGUGG--------------CAAG ........((((...........(((((((.....))((((((((((........)))))))))))))))(((((.....)))))))))(((.(....).))--------------)... ( -30.00) >DroSim_CAF1 82099 106 + 1 GUAAUAACGAAAUAAGAAUUAUAUGCCUUGUGUAGCAAUUCGCUUUAAUAAUCUAUGAAGUGGAUGGGCAGUGGCUUCUUGCCACUUUCGCCUCAAGUGUGG--------------CAAG ........((((...........(((((((.....))((((((((((........)))))))))))))))(((((.....)))))))))(((.(....).))--------------)... ( -30.00) >DroEre_CAF1 86057 120 + 1 GUAAUAACGAAAUAAGAAUUAUAUGCCUUGUGUAGCAAUUCGCUUUAAUAAUCUAUGAAGUGGAUGGGCAGUGGCUUCUUGCCACUUUCGCCUCAAGUGUGGCCAGUGGCAAGUGGCAAG .......................(((((((.....))((((((((((........)))))))))))))))...(((.(((((((((...(((.(....).))).))))))))).)))... ( -38.30) >DroYak_CAF1 71518 120 + 1 GUAAUAACGAAAUAAGAAUUAUAUGCCUUGUGUAGCAAUUCGCUUUAAUAAUCUAUGAAGUGGAUGGGCAGUGGCUUCUUGCCACUUUCGCCUCAAGUGUGGCAAGUGGCAAGUGGCAAG .......................(((((((.....))((((((((((........)))))))))))))))...(((.((((((((((..(((.(....).))))))))))))).)))... ( -39.30) >consensus GUAAUAACGAAAUAAGAAUUAUAUGCCUUGUGUAGCAAUUCGCUUUAAUAAUCUAUGAAGUGGAUGGGCAGUGGCUUCUUGCCACUUUCGCCUCAAGUGUGG______________CAAG ........................((((((.......((((((((((........))))))))))((((((((((.....))))))...)))))))).)).................... (-26.30 = -26.30 + 0.00)

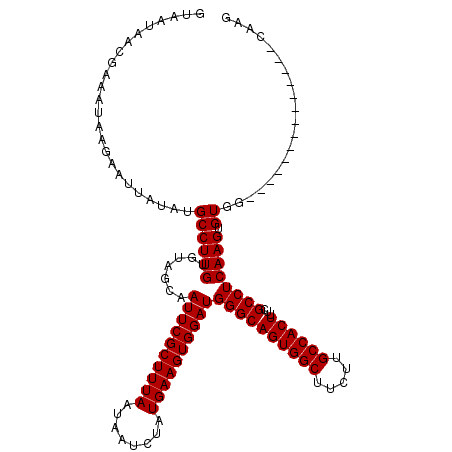

| Location | 16,020,294 – 16,020,400 |
|---|---|
| Length | 106 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.31 |
| Mean single sequence MFE | -24.26 |
| Consensus MFE | -20.20 |
| Energy contribution | -20.20 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.32 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.10 |
| SVM RNA-class probability | 0.585711 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16020294 106 - 22224390 CUUG--------------CCACACUUGAGGCGAAAGUGGCAAGAAGCCACUGCCCAUCCACUUCAUAGAUUAUUAAAGCGAAUUGCUACACAAGGCAUAUAAUUCUUAUUUCGUUAGUAC ..((--------------((.....((.(((...((((((.....)))))))))))....................(((.....)))......))))....................... ( -21.80) >DroSec_CAF1 88046 106 - 1 CUUG--------------CCACACUUGAGGCGAAAGUGGCAAGAAGCCACUGCCCAUCCACUUCAUAGAUUAUUAAAGCGAAUUGCUACACAAGGCAUAUAAUUCUUAUUUCGUUAUUAC ..((--------------((.....((.(((...((((((.....)))))))))))....................(((.....)))......))))....................... ( -21.80) >DroSim_CAF1 82099 106 - 1 CUUG--------------CCACACUUGAGGCGAAAGUGGCAAGAAGCCACUGCCCAUCCACUUCAUAGAUUAUUAAAGCGAAUUGCUACACAAGGCAUAUAAUUCUUAUUUCGUUAUUAC ..((--------------((.....((.(((...((((((.....)))))))))))....................(((.....)))......))))....................... ( -21.80) >DroEre_CAF1 86057 120 - 1 CUUGCCACUUGCCACUGGCCACACUUGAGGCGAAAGUGGCAAGAAGCCACUGCCCAUCCACUUCAUAGAUUAUUAAAGCGAAUUGCUACACAAGGCAUAUAAUUCUUAUUUCGUUAUUAC ..((((.(((((((((.(((........)))...))))))))).................................(((.....)))......))))....................... ( -28.00) >DroYak_CAF1 71518 120 - 1 CUUGCCACUUGCCACUUGCCACACUUGAGGCGAAAGUGGCAAGAAGCCACUGCCCAUCCACUUCAUAGAUUAUUAAAGCGAAUUGCUACACAAGGCAUAUAAUUCUUAUUUCGUUAUUAC ..((((.(((((((((((((........)))..)))))))))).................................(((.....)))......))))....................... ( -27.90) >consensus CUUG______________CCACACUUGAGGCGAAAGUGGCAAGAAGCCACUGCCCAUCCACUUCAUAGAUUAUUAAAGCGAAUUGCUACACAAGGCAUAUAAUUCUUAUUUCGUUAUUAC ............................((((((((((((.....)))))((((......((....))........(((.....)))......))))...........)))))))..... (-20.20 = -20.20 + -0.00)

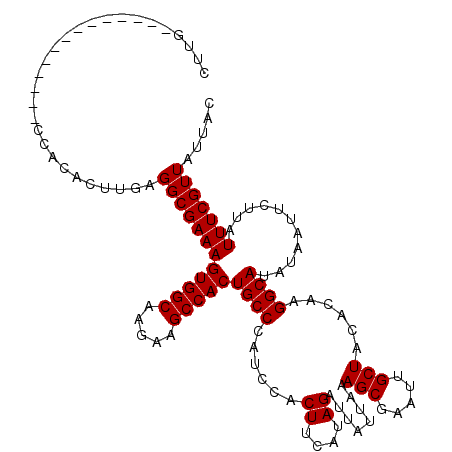

| Location | 16,020,334 – 16,020,434 |
|---|---|
| Length | 100 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 85.69 |
| Mean single sequence MFE | -36.38 |
| Consensus MFE | -27.76 |
| Energy contribution | -27.52 |
| Covariance contribution | -0.24 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.72 |
| Structure conservation index | 0.76 |
| SVM decision value | 1.95 |
| SVM RNA-class probability | 0.983720 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16020334 100 + 22224390 CGCUUUAAUAAUCUAUGAAGUGGAUGGGCAGUGGCUUCUUGCCACUUUCGCCUCAAGUGUGG--------------CAAGUGGCAAGAACUUUGUUAUACUCCCCC------UUUACUGC ..................((((((.(((.((((((((((((((((((..(((.(....).))--------------)))))))))))))....))..)))).))).------)))))).. ( -34.20) >DroSec_CAF1 88086 102 + 1 CGCUUUAAUAAUCUAUGAAGUGGAUGGGCAGUGGCUUCUUGCCACUUUCGCCUCAAGUGUGG--------------CAAGUGGCAAGAACUUUGUUAUACUCCCCCCC----UUAACUGC .((((((........))))))((..(((.((((((((((((((((((..(((.(....).))--------------)))))))))))))....))..)))))))..))----........ ( -33.80) >DroSim_CAF1 82139 101 + 1 CGCUUUAAUAAUCUAUGAAGUGGAUGGGCAGUGGCUUCUUGCCACUUUCGCCUCAAGUGUGG--------------CAAGUGGCAAGAACUUUGUUAUACUCCCCCCC-----UAACUGC .((((((........))))))((..(((.((((((((((((((((((..(((.(....).))--------------)))))))))))))....))..)))))))..))-----....... ( -33.80) >DroEre_CAF1 86097 120 + 1 CGCUUUAAUAAUCUAUGAAGUGGAUGGGCAGUGGCUUCUUGCCACUUUCGCCUCAAGUGUGGCCAGUGGCAAGUGGCAAGUGGCAAGAACUUUGUUAUACUCCUGCACAUUUUUUCCCUC (((((((........)))))))((((.((((.((((.(((((((((...(((.(....).))).))))))))).)))..((((((((...))))))))...))))).))))......... ( -40.60) >DroYak_CAF1 71558 120 + 1 CGCUUUAAUAAUCUAUGAAGUGGAUGGGCAGUGGCUUCUUGCCACUUUCGCCUCAAGUGUGGCAAGUGGCAAGUGGCAAGUGGCAAGAACUUUGUUAUACUCCUGCAAAAUUUUUUCCGC ((((......((((((...))))))....))))(((.((((((((((..(((.(....).))))))))))))).)))..((((.(((((.(((((.........))))).))))).)))) ( -39.50) >consensus CGCUUUAAUAAUCUAUGAAGUGGAUGGGCAGUGGCUUCUUGCCACUUUCGCCUCAAGUGUGG______________CAAGUGGCAAGAACUUUGUUAUACUCCCCCAC____UUUACUGC (((((((........)))))))...(((.(((((((((((((((((((((((....).))))...............))))))))))))....))..)))))))................ (-27.76 = -27.52 + -0.24)

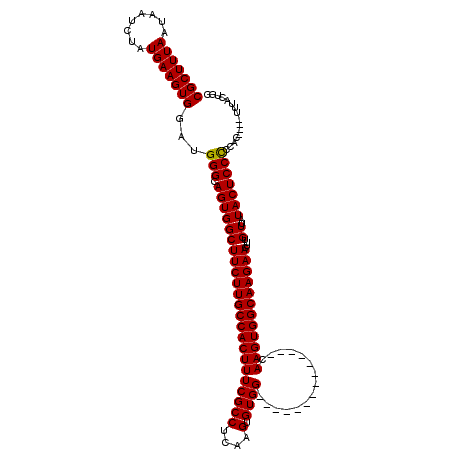

| Location | 16,020,334 – 16,020,434 |
|---|---|
| Length | 100 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 85.69 |
| Mean single sequence MFE | -32.30 |
| Consensus MFE | -20.99 |
| Energy contribution | -20.99 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.99 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.13 |
| SVM RNA-class probability | 0.596018 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16020334 100 - 22224390 GCAGUAAA------GGGGGAGUAUAACAAAGUUCUUGCCACUUG--------------CCACACUUGAGGCGAAAGUGGCAAGAAGCCACUGCCCAUCCACUUCAUAGAUUAUUAAAGCG ((..(((.------.(((.(((.........(((((((((((((--------------((........)))..))))))))))))...))).))).....((....))....)))..)). ( -29.60) >DroSec_CAF1 88086 102 - 1 GCAGUUAA----GGGGGGGAGUAUAACAAAGUUCUUGCCACUUG--------------CCACACUUGAGGCGAAAGUGGCAAGAAGCCACUGCCCAUCCACUUCAUAGAUUAUUAAAGCG ((......----((((((.(((.........(((((((((((((--------------((........)))..))))))))))))...))).))).))).((....)).........)). ( -32.80) >DroSim_CAF1 82139 101 - 1 GCAGUUA-----GGGGGGGAGUAUAACAAAGUUCUUGCCACUUG--------------CCACACUUGAGGCGAAAGUGGCAAGAAGCCACUGCCCAUCCACUUCAUAGAUUAUUAAAGCG ((.....-----((((((.(((.........(((((((((((((--------------((........)))..))))))))))))...))).))).))).((....)).........)). ( -32.80) >DroEre_CAF1 86097 120 - 1 GAGGGAAAAAAUGUGCAGGAGUAUAACAAAGUUCUUGCCACUUGCCACUUGCCACUGGCCACACUUGAGGCGAAAGUGGCAAGAAGCCACUGCCCAUCCACUUCAUAGAUUAUUAAAGCG ((((......(((.((((..((.........(((((((((((((((....(((...)))((....)).)))..))))))))))))))..)))).)))...))))................ ( -34.00) >DroYak_CAF1 71558 120 - 1 GCGGAAAAAAUUUUGCAGGAGUAUAACAAAGUUCUUGCCACUUGCCACUUGCCACUUGCCACACUUGAGGCGAAAGUGGCAAGAAGCCACUGCCCAUCCACUUCAUAGAUUAUUAAAGCG ((((((....)))))).(((((.........((((((((((((((.....))...(((((........)))))))))))))))))((....))......)))))................ ( -32.30) >consensus GCAGUAAA____GGGGGGGAGUAUAACAAAGUUCUUGCCACUUG______________CCACACUUGAGGCGAAAGUGGCAAGAAGCCACUGCCCAUCCACUUCAUAGAUUAUUAAAGCG .................(((((.........((((((((((((..............................))))))))))))((....))......)))))................ (-20.99 = -20.99 + 0.00)


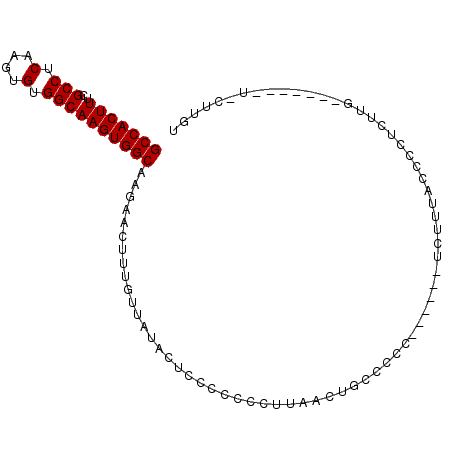
| Location | 16,020,374 – 16,020,473 |
|---|---|
| Length | 99 |
| Sequences | 3 |
| Columns | 101 |
| Reading direction | forward |
| Mean pairwise identity | 83.39 |
| Mean single sequence MFE | -16.11 |
| Consensus MFE | -15.60 |
| Energy contribution | -15.60 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.44 |
| Structure conservation index | 0.97 |
| SVM decision value | 1.74 |
| SVM RNA-class probability | 0.975120 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16020374 99 + 22224390 GCCACUUUCGCCUCAAGUGUGGCAAGUGGCAAGAACUUUGUUAUACUCCCCC--UUUACUGCCCCCCCCCCUUUUUUUCCGCUCUUUCUGUUCGUCCCAGU (((((((..(((.(....).))))))))))..((((................--...................................))))........ ( -16.74) >DroSec_CAF1 88126 87 + 1 GCCACUUUCGCCUCAAGUGUGGCAAGUGGCAAGAACUUUGUUAUACUCCCCCCCUUAACUGCCCCC------UCUUUACCCCUCUUG-------U-CUUGU (((((((..(((.(....).))))))))))((((.....((((............)))).......------))))...........-------.-..... ( -16.00) >DroSim_CAF1 82179 86 + 1 GCCACUUUCGCCUCAAGUGUGGCAAGUGGCAAGAACUUUGUUAUACUCCCCCCC-UAACUGCCCCC------UCUUUACCCCUCUUG-------U-CUUGU (((((((..(((.(....).))))))))))........................-...........------...............-------.-..... ( -15.60) >consensus GCCACUUUCGCCUCAAGUGUGGCAAGUGGCAAGAACUUUGUUAUACUCCCCCCCUUAACUGCCCCC______UCUUUACCCCUCUUG_______U_CUUGU (((((((..(((.(....).))))))))))....................................................................... (-15.60 = -15.60 + -0.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:12:01 2006