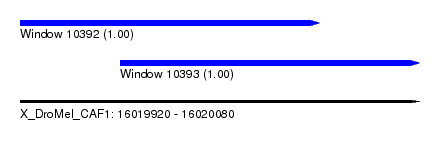
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 16,019,920 – 16,020,080 |
| Length | 160 |
| Max. P | 0.997895 |

| Location | 16,019,920 – 16,020,040 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 100.00 |
| Mean single sequence MFE | -27.30 |
| Consensus MFE | -27.30 |
| Energy contribution | -27.30 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.21 |
| Structure conservation index | 1.00 |
| SVM decision value | 2.68 |
| SVM RNA-class probability | 0.996328 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
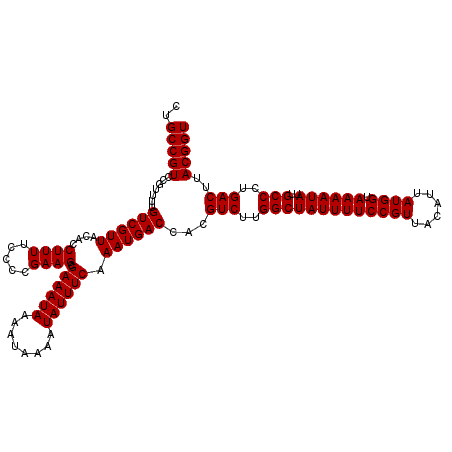
>X_DroMel_CAF1 16019920 120 + 22224390 CUGCCGUCCGUUUUGUCGUUACACCUUUUCCCCGAAGCGAAAUAAAAUAAAAUAUUUCAAAUGACCACGUCUUGGCUAUUUUCCGUUACAUUAUGGUAAAAUAUUGCCCUGACUUACGGU ..(((((.......((((((....((((.....)))).((((((........)))))).))))))...(((..(((((((((((((......)))).))))))..)))..)))..))))) ( -27.30) >DroSec_CAF1 87676 120 + 1 CUGCCGUCCGUUUUGUCGUUACACCUUUUCCCCGAAGCGAAAUAAAAUAAAAUAUUUCAAAUGACCACGUCUUGGCUAUUUUCCGUUACAUUAUGGUAAAAUAUUGCCCUGACUUACGGU ..(((((.......((((((....((((.....)))).((((((........)))))).))))))...(((..(((((((((((((......)))).))))))..)))..)))..))))) ( -27.30) >DroSim_CAF1 81729 120 + 1 CUGCCGUCCGUUUUGUCGUUACACCUUUUCCCCGAAGCGAAAUAAAAUAAAAUAUUUCAAAUGACCACGUCUUGGCUAUUUUCCGUUACAUUAUGGUAAAAUAUUGCCCUGACUUACGGU ..(((((.......((((((....((((.....)))).((((((........)))))).))))))...(((..(((((((((((((......)))).))))))..)))..)))..))))) ( -27.30) >DroEre_CAF1 85679 120 + 1 CUGCCGUCCGUUUUGUCGUUACACCUUUUCCCCGAAGCGAAAUAAAAUAAAAUAUUUCAAAUGACCACGUCUUGGCUAUUUUCCGUUACAUUAUGGUAAAAUAUUGCCCUGACUUACGGU ..(((((.......((((((....((((.....)))).((((((........)))))).))))))...(((..(((((((((((((......)))).))))))..)))..)))..))))) ( -27.30) >DroYak_CAF1 71128 120 + 1 CUGCCGUCCGUUUUGUCGUUACACCUUUUCCCCGAAGCGAAAUAAAAUAAAAUAUUUCAAAUGACCACGUCUUGGCUAUUUUCCGUUACAUUAUGGUAAAAUAUUGCCCUGACUUACGGU ..(((((.......((((((....((((.....)))).((((((........)))))).))))))...(((..(((((((((((((......)))).))))))..)))..)))..))))) ( -27.30) >consensus CUGCCGUCCGUUUUGUCGUUACACCUUUUCCCCGAAGCGAAAUAAAAUAAAAUAUUUCAAAUGACCACGUCUUGGCUAUUUUCCGUUACAUUAUGGUAAAAUAUUGCCCUGACUUACGGU ..(((((.......((((((....((((.....)))).((((((........)))))).))))))...(((..(((((((((((((......)))).))))))..)))..)))..))))) (-27.30 = -27.30 + 0.00)

| Location | 16,019,960 – 16,020,080 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.26 |
| Mean single sequence MFE | -28.14 |
| Consensus MFE | -25.54 |
| Energy contribution | -25.94 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.08 |
| Structure conservation index | 0.91 |
| SVM decision value | 2.96 |
| SVM RNA-class probability | 0.997895 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16019960 120 + 22224390 AAUAAAAUAAAAUAUUUCAAAUGACCACGUCUUGGCUAUUUUCCGUUACAUUAUGGUAAAAUAUUGCCCUGACUUACGGUCGAUACAGCCGUCGUCUCGGUUUUCGACUUGGGUUUCAGU ......................((((..(((..(((((((((((((......)))).))))))..)))..)))....((((((...(((((......))))).))))))..))))..... ( -30.80) >DroSec_CAF1 87716 117 + 1 AAUAAAAUAAAAUAUUUCAAAUGACCACGUCUUGGCUAUUUUCCGUUACAUUAUGGUAAAAUAUUGCCCUGACUUACGGUCGAUACAGCCG---UCUCGGUUUUCGACUUGGGUUUCAGU ......................((((..(((..(((((((((((((......)))).))))))..)))..)))....((((((...(((((---...))))).))))))..))))..... ( -29.00) >DroSim_CAF1 81769 117 + 1 AAUAAAAUAAAAUAUUUCAAAUGACCACGUCUUGGCUAUUUUCCGUUACAUUAUGGUAAAAUAUUGCCCUGACUUACGGUCGAUACAGCCG---UCUCGGUUUUCGACUUGGGUUUCAGU ......................((((..(((..(((((((((((((......)))).))))))..)))..)))....((((((...(((((---...))))).))))))..))))..... ( -29.00) >DroEre_CAF1 85719 109 + 1 AAUAAAAUAAAAUAUUUCAAAUGACCACGUCUUGGCUAUUUUCCGUUACAUUAUGGUAAAAUAUUGCCCUGACUUACGGUCGAUACAGCCG---UCUCGGUUUUCUACAUGU-------- ......................((((..(((..(((((((((((((......)))).))))))..)))..)))....))))((...(((((---...))))).)).......-------- ( -22.90) >DroYak_CAF1 71168 117 + 1 AAUAAAAUAAAAUAUUUCAAAUGACCACGUCUUGGCUAUUUUCCGUUACAUUAUGGUAAAAUAUUGCCCUGACUUACGGUCGAUACAGCCG---UCUCGGUUUUCGACUUGGGUUUCAGU ......................((((..(((..(((((((((((((......)))).))))))..)))..)))....((((((...(((((---...))))).))))))..))))..... ( -29.00) >consensus AAUAAAAUAAAAUAUUUCAAAUGACCACGUCUUGGCUAUUUUCCGUUACAUUAUGGUAAAAUAUUGCCCUGACUUACGGUCGAUACAGCCG___UCUCGGUUUUCGACUUGGGUUUCAGU ........................(((.(((..(((((((((((((......)))).))))))..)))..))).....(((((...(((((......))))).))))).)))........ (-25.54 = -25.94 + 0.40)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:11:55 2006