
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 16,017,128 – 16,017,387 |
| Length | 259 |
| Max. P | 0.999974 |

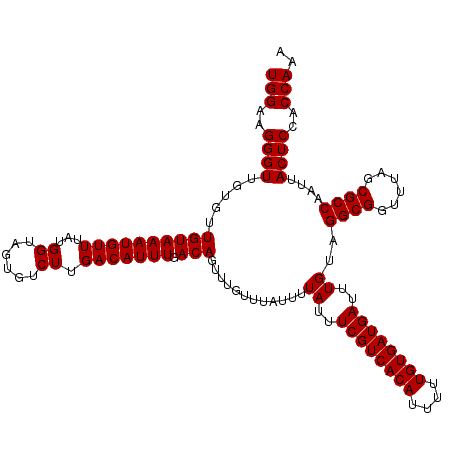
| Location | 16,017,128 – 16,017,248 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 97.15 |
| Mean single sequence MFE | -30.60 |
| Consensus MFE | -28.24 |
| Energy contribution | -28.24 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.20 |
| Structure conservation index | 0.92 |
| SVM decision value | 3.05 |
| SVM RNA-class probability | 0.998257 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16017128 120 + 22224390 UGGAAGGGUUGUGUUGUAAAUGUUUAUGGUAGUGUCUUGACAUUUGACAGUUUGUUUAUUUUAUUUCGUCACAUUUUUGUGAUGAUUUGUAGGCGGUUUAGCGCCAAUUACUCCACCAAA ((((..((((((((((((((((((...((......)).)))))))).....((((((((......((((((((....))))))))...))))))))..))))).)))))..))))..... ( -30.60) >DroSec_CAF1 84977 120 + 1 UGGAAGGGUUGUGUUGUAAAUGUUUAUGGUAGUGUCUUGACAUUUGACAGUUUGUUUAUUUUAUUUCGUCACAUUUUUGUGAUGAUUUGUAGGCGGUUUAGCGCCAAUUACUCCACCAAA ((((..((((((((((((((((((...((......)).)))))))).....((((((((......((((((((....))))))))...))))))))..))))).)))))..))))..... ( -30.60) >DroSim_CAF1 78991 120 + 1 UGGAAGGGUUGUGUUGUAAAUGUUUAUGGUAGUGUCUUGACAUUUGACAGUUUGUUUAUUUUAUUUCGUCACAUUUUUGUGAUGAUUUGUAGGCGGUUUAGCGCCAAUUACUCCACCAAA ((((..((((((((((((((((((...((......)).)))))))).....((((((((......((((((((....))))))))...))))))))..))))).)))))..))))..... ( -30.60) >DroEre_CAF1 83043 115 + 1 UGGAGGGGU-----UGUAAAUGUUUAUGGUAGUGUCUUGACAUUUGACAGUUUGUUUAUUUUAUUUCGUCACAUUUUUGUGAUGAUUUGUAGGCGGUUUAGCGCCAAUUACUCCACCAAA (((.(((((-----((((((((((...((......)).))))))).)))............((..((((((((....))))))))..))..((((......))))....))))).))).. ( -32.70) >DroYak_CAF1 68228 115 + 1 UGGAAGGGU-----UGUAAAUGUUUAUGGUAGUGUCUUGACAUUUGACAGUUUGUUUAUUUUAUUUCGUCACAUUUUUGUGAUGAUUUGUAGGCGGUUUAGCGCCAAUUACUCCACCAAA (((..((((-----((((((((((...((......)).))))))).)))............((..((((((((....))))))))..))..((((......))))....))))..))).. ( -28.50) >consensus UGGAAGGGUUGUGUUGUAAAUGUUUAUGGUAGUGUCUUGACAUUUGACAGUUUGUUUAUUUUAUUUCGUCACAUUUUUGUGAUGAUUUGUAGGCGGUUUAGCGCCAAUUACUCCACCAAA (((..((((.....((((((((((...((......)).))))))).)))............((..((((((((....))))))))..))..((((......))))....))))..))).. (-28.24 = -28.24 + 0.00)


| Location | 16,017,128 – 16,017,248 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.15 |
| Mean single sequence MFE | -21.20 |
| Consensus MFE | -21.20 |
| Energy contribution | -21.20 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.95 |
| Structure conservation index | 1.00 |
| SVM decision value | 5.10 |
| SVM RNA-class probability | 0.999974 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16017128 120 - 22224390 UUUGGUGGAGUAAUUGGCGCUAAACCGCCUACAAAUCAUCACAAAAAUGUGACGAAAUAAAAUAAACAAACUGUCAAAUGUCAAGACACUACCAUAAACAUUUACAACACAACCCUUCCA ..(((((((((....((((......))))......((.(((((....))))).))..............)))(((.........))).)))))).......................... ( -21.20) >DroSec_CAF1 84977 120 - 1 UUUGGUGGAGUAAUUGGCGCUAAACCGCCUACAAAUCAUCACAAAAAUGUGACGAAAUAAAAUAAACAAACUGUCAAAUGUCAAGACACUACCAUAAACAUUUACAACACAACCCUUCCA ..(((((((((....((((......))))......((.(((((....))))).))..............)))(((.........))).)))))).......................... ( -21.20) >DroSim_CAF1 78991 120 - 1 UUUGGUGGAGUAAUUGGCGCUAAACCGCCUACAAAUCAUCACAAAAAUGUGACGAAAUAAAAUAAACAAACUGUCAAAUGUCAAGACACUACCAUAAACAUUUACAACACAACCCUUCCA ..(((((((((....((((......))))......((.(((((....))))).))..............)))(((.........))).)))))).......................... ( -21.20) >DroEre_CAF1 83043 115 - 1 UUUGGUGGAGUAAUUGGCGCUAAACCGCCUACAAAUCAUCACAAAAAUGUGACGAAAUAAAAUAAACAAACUGUCAAAUGUCAAGACACUACCAUAAACAUUUACA-----ACCCCUCCA ..(((((((((....((((......))))......((.(((((....))))).))..............)))(((.........))).))))))............-----......... ( -21.20) >DroYak_CAF1 68228 115 - 1 UUUGGUGGAGUAAUUGGCGCUAAACCGCCUACAAAUCAUCACAAAAAUGUGACGAAAUAAAAUAAACAAACUGUCAAAUGUCAAGACACUACCAUAAACAUUUACA-----ACCCUUCCA ..(((((((((....((((......))))......((.(((((....))))).))..............)))(((.........))).))))))............-----......... ( -21.20) >consensus UUUGGUGGAGUAAUUGGCGCUAAACCGCCUACAAAUCAUCACAAAAAUGUGACGAAAUAAAAUAAACAAACUGUCAAAUGUCAAGACACUACCAUAAACAUUUACAACACAACCCUUCCA ..(((((((((....((((......))))......((.(((((....))))).))..............)))(((.........))).)))))).......................... (-21.20 = -21.20 + -0.00)

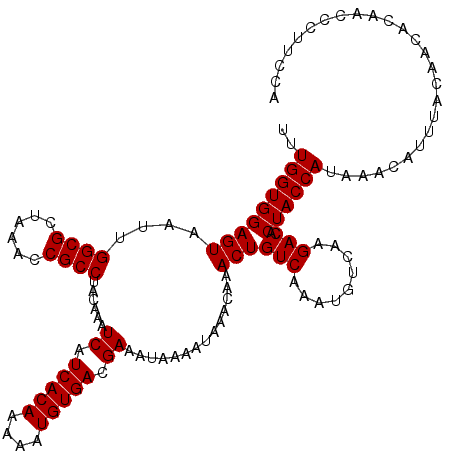
| Location | 16,017,168 – 16,017,288 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 97.83 |
| Mean single sequence MFE | -29.26 |
| Consensus MFE | -25.32 |
| Energy contribution | -26.56 |
| Covariance contribution | 1.24 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.28 |
| Structure conservation index | 0.87 |
| SVM decision value | 2.01 |
| SVM RNA-class probability | 0.985540 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16017168 120 + 22224390 CAUUUGACAGUUUGUUUAUUUUAUUUCGUCACAUUUUUGUGAUGAUUUGUAGGCGGUUUAGCGCCAAUUACUCCACCAAACUUUUACAAUGCAGACUCUGGUAAACGGCAGUCUUUAAAA (((((((.((((((.......((..((((((((....))))))))..))..((((......))))...........)))))).))).)))).((((((((.....))).)))))...... ( -31.50) >DroSec_CAF1 85017 120 + 1 CAUUUGACAGUUUGUUUAUUUUAUUUCGUCACAUUUUUGUGAUGAUUUGUAGGCGGUUUAGCGCCAAUUACUCCACCAAACUUUUACAAUGCAGUCUCUGGUAAACGGCAGUCUUUAAAA (((((((.((((((.......((..((((((((....))))))))..))..((((......))))...........)))))).))).)))).((.(((((.....))).)).))...... ( -27.60) >DroSim_CAF1 79031 120 + 1 CAUUUGACAGUUUGUUUAUUUUAUUUCGUCACAUUUUUGUGAUGAUUUGUAGGCGGUUUAGCGCCAAUUACUCCACCAAACUUUUACAAUGCAGUCUCUGGUAAACGGCAGUCUUUAAAA (((((((.((((((.......((..((((((((....))))))))..))..((((......))))...........)))))).))).)))).((.(((((.....))).)).))...... ( -27.60) >DroEre_CAF1 83078 118 + 1 CAUUUGACAGUUUGUUUAUUUUAUUUCGUCACAUUUUUGUGAUGAUUUGUAGGCGGUUUAGCGCCAAUUACUCCACCAAACUUUUACGCUGCAGAC--UGGUAAACGGCAGUCUUUAAAA ....(((.((((((.......((..((((((((....))))))))..))..((((......))))...........)))))).)))......((((--((........))))))...... ( -29.10) >DroYak_CAF1 68263 118 + 1 CAUUUGACAGUUUGUUUAUUUUAUUUCGUCACAUUUUUGUGAUGAUUUGUAGGCGGUUUAGCGCCAAUUACUCCACCAAACUUUUACAAUGCAGAC--UGGUAAACGGCAGUCUUUAAAA (((((((.((((((.......((..((((((((....))))))))..))..((((......))))...........)))))).))).)))).((((--((........))))))...... ( -30.50) >consensus CAUUUGACAGUUUGUUUAUUUUAUUUCGUCACAUUUUUGUGAUGAUUUGUAGGCGGUUUAGCGCCAAUUACUCCACCAAACUUUUACAAUGCAGACUCUGGUAAACGGCAGUCUUUAAAA (((((((.((((((.......((..((((((((....))))))))..))..((((......))))...........)))))).))).)))).((((((((.....))).)))))...... (-25.32 = -26.56 + 1.24)

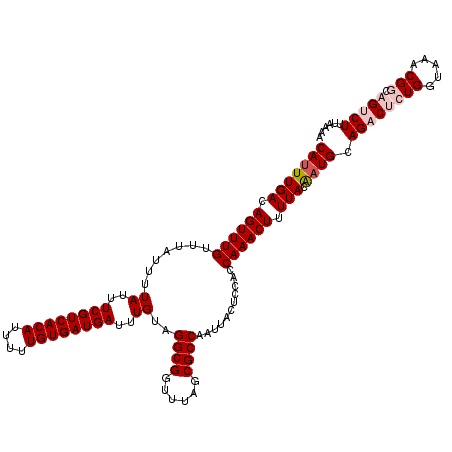
| Location | 16,017,168 – 16,017,288 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.83 |
| Mean single sequence MFE | -24.64 |
| Consensus MFE | -20.90 |
| Energy contribution | -20.90 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.53 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.13 |
| SVM RNA-class probability | 0.597087 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16017168 120 - 22224390 UUUUAAAGACUGCCGUUUACCAGAGUCUGCAUUGUAAAAGUUUGGUGGAGUAAUUGGCGCUAAACCGCCUACAAAUCAUCACAAAAAUGUGACGAAAUAAAAUAAACAAACUGUCAAAUG ......(((((............))))).((((.....((((((.....(((...((((......)))))))...((.(((((....))))).))...........))))))....)))) ( -22.40) >DroSec_CAF1 85017 120 - 1 UUUUAAAGACUGCCGUUUACCAGAGACUGCAUUGUAAAAGUUUGGUGGAGUAAUUGGCGCUAAACCGCCUACAAAUCAUCACAAAAAUGUGACGAAAUAAAAUAAACAAACUGUCAAAUG .......((((((.((((.....)))).))).......((((((.....(((...((((......)))))))...((.(((((....))))).))...........)))))))))..... ( -24.90) >DroSim_CAF1 79031 120 - 1 UUUUAAAGACUGCCGUUUACCAGAGACUGCAUUGUAAAAGUUUGGUGGAGUAAUUGGCGCUAAACCGCCUACAAAUCAUCACAAAAAUGUGACGAAAUAAAAUAAACAAACUGUCAAAUG .......((((((.((((.....)))).))).......((((((.....(((...((((......)))))))...((.(((((....))))).))...........)))))))))..... ( -24.90) >DroEre_CAF1 83078 118 - 1 UUUUAAAGACUGCCGUUUACCA--GUCUGCAGCGUAAAAGUUUGGUGGAGUAAUUGGCGCUAAACCGCCUACAAAUCAUCACAAAAAUGUGACGAAAUAAAAUAAACAAACUGUCAAAUG ......((((((........))--))))...........((((((..(.(((...((((......)))))))...((.(((((....))))).))...............)..)))))). ( -25.40) >DroYak_CAF1 68263 118 - 1 UUUUAAAGACUGCCGUUUACCA--GUCUGCAUUGUAAAAGUUUGGUGGAGUAAUUGGCGCUAAACCGCCUACAAAUCAUCACAAAAAUGUGACGAAAUAAAAUAAACAAACUGUCAAAUG ......((((((........))--)))).((((.....((((((.....(((...((((......)))))))...((.(((((....))))).))...........))))))....)))) ( -25.60) >consensus UUUUAAAGACUGCCGUUUACCAGAGUCUGCAUUGUAAAAGUUUGGUGGAGUAAUUGGCGCUAAACCGCCUACAAAUCAUCACAAAAAUGUGACGAAAUAAAAUAAACAAACUGUCAAAUG .......((((((.(...........).))).......((((((.....(((...((((......)))))))...((.(((((....))))).))...........)))))))))..... (-20.90 = -20.90 + 0.00)

| Location | 16,017,288 – 16,017,387 |
|---|---|
| Length | 99 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 72.57 |
| Mean single sequence MFE | -25.87 |
| Consensus MFE | -12.91 |
| Energy contribution | -13.47 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.83 |
| Structure conservation index | 0.50 |
| SVM decision value | 0.62 |
| SVM RNA-class probability | 0.801309 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16017288 99 + 22224390 AUAACACAUUUGGCGCCAAUUACCAA-AUGACCGAGCCAACGCCGAAUCUUCUCAAAUGUCAGGGGCUA-AAUAUCGCUAACUUAAGCUAUUUGGUUAAAA------------------- ......(((((((.........))))-))).(((.((....)))((......))........))(((((-((((..(((......))))))))))))....------------------- ( -17.00) >DroSec_CAF1 85137 117 + 1 AUAACACAUUUGGCGCCAAUUACCAA-AUGACCGAGCCACCGACGAAUGUUCUCAAAUGUCAGGGGCUA-AAUAUCGCUAGCUUCAGCUAGCUGGUUAAUAUAGGUCAAAUAUGGAGUA- ....((.(((((((.......((((.-.......((((.((((((..((....))..)))).)))))).-......((((((....))))))))))........))))))).)).....- ( -33.06) >DroSim_CAF1 79151 118 + 1 AUAACACAUUUGGCGCCAAUUACCAAAAAGACCGAGCCACCGACGAAUGCUCUCAAAUGUCAGGGGCUA-AAUAUCGCUAGCUUCAGCUAGCUGGUUAAUAUAGGUUAAAUAUGGAGUA- ............((.(((...(((.....(((((((((.((((((..((....))..)))).)))))).-......((((((....)))))))))))......)))......))).)).- ( -30.60) >DroEre_CAF1 83196 76 + 1 AUAACGCAUUUGGCGCCAAUU--------------------GCCGAAUGUUCUUAAAUGCCAGGGCAUA-CAUAUCGCUAGCUUCAGCUAGCUGGCU----------------------- .....((((((((((.....)--------------------)))))))))......((((....)))).-......(((((((......))))))).----------------------- ( -27.30) >DroYak_CAF1 68381 113 + 1 AUAACACAUUUGGCGCCAAUUACCAA-AUGACCUAGCCACCGCCGAAUGUUCUCAAAUGUCAGGGUACAAAAUAUCGCUACCUUUAGCUAGCUAGUUCAUUUUC------UUUGGAAUAA ...............((((.....((-((((.(((((..........(((.(((........))).))).......((((....))))..))))).))))))..------.))))..... ( -21.40) >consensus AUAACACAUUUGGCGCCAAUUACCAA_AUGACCGAGCCACCGCCGAAUGUUCUCAAAUGUCAGGGGCUA_AAUAUCGCUAGCUUCAGCUAGCUGGUUAAUAU________U_UGGA_UA_ .....((((((((((.........................)))))))))).(((........)))...........(((((((......)))))))........................ (-12.91 = -13.47 + 0.56)


| Location | 16,017,288 – 16,017,387 |
|---|---|
| Length | 99 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 72.57 |
| Mean single sequence MFE | -29.26 |
| Consensus MFE | -12.03 |
| Energy contribution | -13.34 |
| Covariance contribution | 1.31 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.81 |
| Structure conservation index | 0.41 |
| SVM decision value | 0.39 |
| SVM RNA-class probability | 0.715466 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16017288 99 - 22224390 -------------------UUUUAACCAAAUAGCUUAAGUUAGCGAUAUU-UAGCCCCUGACAUUUGAGAAGAUUCGGCGUUGGCUCGGUCAU-UUGGUAAUUGGCGCCAAAUGUGUUAU -------------------.....(((((((.(((......)))(((...-.(((((((((.((((....)))))))).)..))))..)))))-)))))...((((((.....)))))). ( -22.90) >DroSec_CAF1 85137 117 - 1 -UACUCCAUAUUUGACCUAUAUUAACCAGCUAGCUGAAGCUAGCGAUAUU-UAGCCCCUGACAUUUGAGAACAUUCGUCGGUGGCUCGGUCAU-UUGGUAAUUGGCGCCAAAUGUGUUAU -.....((((((((.((((.((((.(((((((((....))))))(((...-.((((.(((((...((....))...))))).))))..)))..-.)))))))))).).)))))))).... ( -37.50) >DroSim_CAF1 79151 118 - 1 -UACUCCAUAUUUAACCUAUAUUAACCAGCUAGCUGAAGCUAGCGAUAUU-UAGCCCCUGACAUUUGAGAGCAUUCGUCGGUGGCUCGGUCUUUUUGGUAAUUGGCGCCAAAUGUGUUAU -.....(((((((..((((.((((.(((((((((....))))))(((...-.((((.(((((...((....))...))))).))))..)))....)))))))))).)..))))))).... ( -33.60) >DroEre_CAF1 83196 76 - 1 -----------------------AGCCAGCUAGCUGAAGCUAGCGAUAUG-UAUGCCCUGGCAUUUAAGAACAUUCGGC--------------------AAUUGGCGCCAAAUGCGUUAU -----------------------.(((.((((((....))))))((.(((-(((((....))))......)))))))))--------------------...((((((.....)))))). ( -27.00) >DroYak_CAF1 68381 113 - 1 UUAUUCCAAA------GAAAAUGAACUAGCUAGCUAAAGGUAGCGAUAUUUUGUACCCUGACAUUUGAGAACAUUCGGCGGUGGCUAGGUCAU-UUGGUAAUUGGCGCCAAAUGUGUUAU ..........------..((((((.((((((.((((....))))..........(((((((....((....)).)))).))))))))).))))-))......((((((.....)))))). ( -25.30) >consensus _UA_UCCA_A________AUAUUAACCAGCUAGCUGAAGCUAGCGAUAUU_UAGCCCCUGACAUUUGAGAACAUUCGGCGGUGGCUCGGUCAU_UUGGUAAUUGGCGCCAAAUGUGUUAU ............................((((((....))))))................(((((((.((....))(.((((.(((..........))).)))).)..)))))))..... (-12.03 = -13.34 + 1.31)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:11:51 2006