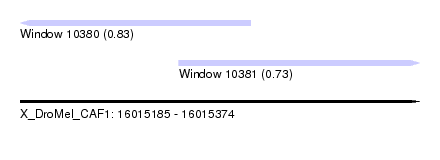
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 16,015,185 – 16,015,374 |
| Length | 189 |
| Max. P | 0.832419 |

| Location | 16,015,185 – 16,015,294 |
|---|---|
| Length | 109 |
| Sequences | 3 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 93.12 |
| Mean single sequence MFE | -34.53 |
| Consensus MFE | -32.33 |
| Energy contribution | -31.67 |
| Covariance contribution | -0.66 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.93 |
| Structure conservation index | 0.94 |
| SVM decision value | 0.72 |
| SVM RNA-class probability | 0.832419 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
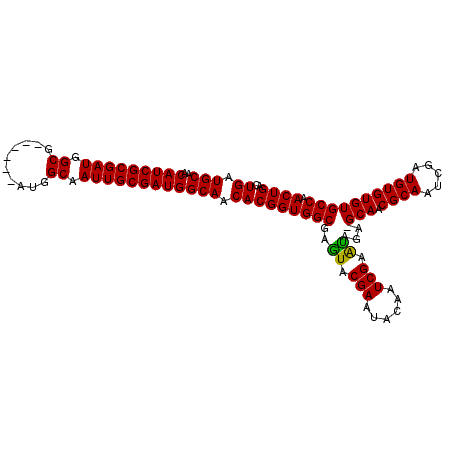
>X_DroMel_CAF1 16015185 109 - 22224390 UGAUGCAACAUCGCGAUGGCGAUGGCGAUGGCGAUUGCGAUGGCAACACGGUGGCGAAUACGAAUACAAUCGAAUAGAGGCAACGCAAUCGAUGUGUGUGCCAACUGAC ((.(((..(((((((((.((.((....)).)).)))))))))))).))((((((((.......(((((.((((.....(....)....))))))))).)))).)))).. ( -37.70) >DroSec_CAF1 83083 102 - 1 UGAUGCAACAUCGCGAUGGCG------AUGGCAAUUGCGAUGGCAACACGGUGGCGAGUACGAAUACAAUCGAAUAGA-GCAACGCAAUCGAUGUGUGUGCCAACUGAC ((.(((..(((((((((.((.------...)).)))))))))))).))(((((((.....(((......)))......-(((.((((.....)))))))))).)))).. ( -32.10) >DroSim_CAF1 77077 102 - 1 UGAUGCAACAUCGCGAUGGCG------AUGGCAAUUGCGAUGGCAACACGGUGGCGAGUACGAAUACAAUCGAGCAGA-GCAACGCAAUCGAUGUGUGUGCCAACUGAC ((.(((..(((((((((.((.------...)).)))))))))))).))(((((((..((.(((......))).))...-(((.((((.....)))))))))).)))).. ( -33.80) >consensus UGAUGCAACAUCGCGAUGGCG______AUGGCAAUUGCGAUGGCAACACGGUGGCGAGUACGAAUACAAUCGAAUAGA_GCAACGCAAUCGAUGUGUGUGCCAACUGAC ((.(((..(((((((((.((..........)).)))))))))))).))(((((((..((.(((......))).))....(((.((((.....)))))))))).)))).. (-32.33 = -31.67 + -0.66)


| Location | 16,015,260 – 16,015,374 |
|---|---|
| Length | 114 |
| Sequences | 4 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 86.12 |
| Mean single sequence MFE | -32.50 |
| Consensus MFE | -23.31 |
| Energy contribution | -23.50 |
| Covariance contribution | 0.19 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.80 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.43 |
| SVM RNA-class probability | 0.732783 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16015260 114 + 22224390 UCGCCAUCGCCAUCGCCAUCGCGAUGUUGCAUCAAAUGCGCUGUAAUAUACUAACGUCUUUUGGUGUUUUGCUACGAUUUUCUGGCGAUCCACUUGAGUUGCAGUGGGGCACAA ..(((...((((((((....))))))..))........(((((((((..((....))..((.((((..((((((.(.....)))))))..)))).))))))))))).))).... ( -36.90) >DroSec_CAF1 83157 107 + 1 UUGCCAU------CGCCAUCGCGAUGUUGCAUCAAAUUCGCUGUAAUAUACUAACGUCUUUUGCUGU-UUGCUGCGAUUCUCUGGCGAUCCACUUGAGUUGCAGUGGGGCACAA .((((((------(((((..(.((.((((((.(((((.....((((...((....))...)))).))-))).)))))))).))))))))(((((........)))))))))... ( -34.40) >DroSim_CAF1 77151 107 + 1 UUGCCAU------CGCCAUCGCGAUGUUGCAUCAAAUUCGCUGUAAUAUACUAACGUCUUUUUGUGU-UUGCUGCGAUUUUCUGGCGAUUCACUUGAGUUGUAGUGGGGCACAA .((((((------(((((....((((...))))(((.((((.((((.((((............))))-)))).)))).))).)))))))(((((........)))))))))... ( -31.50) >DroEre_CAF1 81528 93 + 1 ------------------UCGCGAUGUUGCAUCAAAUGCGCUGUAAUAUACUGACGUCU--UGCUGU-UUGCUGCGAUUCUCUGGCGAUCCACUUGAGUUGCAGUGGGGCACAA ------------------....((((((((((...))))...(((...))).)))))).--((((..-(..(((((((((..(((....)))...)))))))))..)))))... ( -27.20) >consensus UUGCCAU______CGCCAUCGCGAUGUUGCAUCAAAUGCGCUGUAAUAUACUAACGUCUUUUGCUGU_UUGCUGCGAUUCUCUGGCGAUCCACUUGAGUUGCAGUGGGGCACAA .......................((((((((.(......).))))))))..............((((.(..(((((((((..(((....)))...)))))))))..).)))).. (-23.31 = -23.50 + 0.19)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:11:44 2006