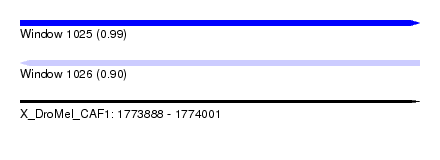
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 1,773,888 – 1,774,001 |
| Length | 113 |
| Max. P | 0.988966 |

| Location | 1,773,888 – 1,774,001 |
|---|---|
| Length | 113 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 77.25 |
| Mean single sequence MFE | -56.42 |
| Consensus MFE | -38.48 |
| Energy contribution | -38.73 |
| Covariance contribution | 0.26 |
| Combinations/Pair | 1.45 |
| Mean z-score | -2.88 |
| Structure conservation index | 0.68 |
| SVM decision value | 2.14 |
| SVM RNA-class probability | 0.988966 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
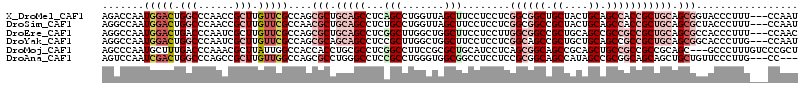
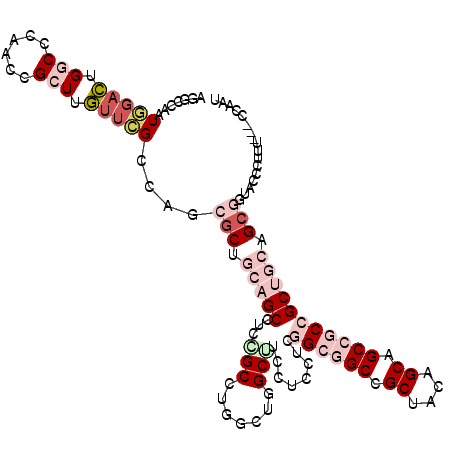
>X_DroMel_CAF1 1773888 113 + 22224390 AUUGG---AAAGGGUACCGCUGCAGCGGUGGCUGCAGUAGCAGCCGCCGAGGAGGAAGCUAACCAGGCUGAGGCUGCAGCGCUGGCGAACAAGCGGUUGGGCCAGUCCAUUGGUCU ..(((---(...(((((((((((((((((((((((....)))))))))........((((.....))))...))))))))(((........))))))...)))..))))....... ( -55.20) >DroSim_CAF1 4628 113 + 1 AUUGG---AAAGGGUAGCGCUGCAGCGGUGGCUGCAGUAGCGGCCGCCGAGGAGGAAGCUAACCAGGCAGAGGCUGCAGCGUUGGCGAACAAGCGGUUGGGCCAGUCCAUUGGCCU .....---....(.(((((((((((((((((((((....))))))))).........(((.....)))....)))))))))))).)............(((((((....))))))) ( -55.10) >DroEre_CAF1 6930 113 + 1 GUUGG---AAAGGGUGGCGCUGCAGCGGCGGCGGCUGCAGCGGCCGCCAAGGAGGAAGCCAGCCAAGCCGAGGCUGCAGCGCUGGCGAACAAGCGAUUGGGUCAGUCCAUUGGCCU (((..---....(.((((((((((((..(((((((((..((..((.(....).))..)))))))..))))..)))))))))))).).....)))....(((((((....))))))) ( -55.00) >DroYak_CAF1 7074 113 + 1 AUUGG---CAAGGGUGCCGCUGCAGCGGCGGCUGCAGCAGCGGCUGCCGAGGAGGAAGCCAGCCAAGCGGAGGCUGCUGCGCUGGCGAACAAGCGAUUGGGCCAGUCCAUUGGCCU ...((---((....))))((((((((....)))))))).((((((.((.....)).)))).((((.(((.((....)).)))))))......))....(((((((....))))))) ( -56.90) >DroMoj_CAF1 12034 113 + 1 AGCGGGACAAAGGGC---GCUGCGGCGGCGGCAGCUGCGGCUGCCGCUGAGGAUGCAGCGCGGAAGGCCGAGGCGCAGGUGGUGGCCAAUAAGCGUUUGGGUCAAAGCAUUGGGCU .((..........((---((((((.((((((((((....))))))))))....))))))))....((((.((((((...(((...)))....)))))).))))...))........ ( -57.50) >DroAna_CAF1 7172 110 + 1 ---GG---CAAGGGAACAGCAGCUGCUGCCGCGGCUAUGGCUGCCGCGGAGGAGGCCGCCACCCAGGCGGAGGCCCAGGCGCUGGCCAACAAGCGGCUGGGCCAGUCGAUUGGACU ---.(---(..(((....((.(((.((.(((((((.......))))))).)).))).))..)))..))...(((((((.((((........)))).)))))))((((.....)))) ( -58.80) >consensus AUUGG___AAAGGGUACCGCUGCAGCGGCGGCUGCAGCAGCGGCCGCCGAGGAGGAAGCCAGCCAGGCCGAGGCUGCAGCGCUGGCGAACAAGCGGUUGGGCCAGUCCAUUGGCCU ..................(((((((((((((((((....))))))))).........((.......))....))))))))(((........)))....(((((((....))))))) (-38.48 = -38.73 + 0.26)

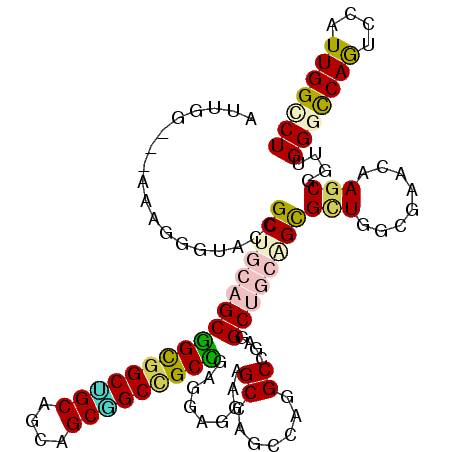
| Location | 1,773,888 – 1,774,001 |
|---|---|
| Length | 113 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 77.25 |
| Mean single sequence MFE | -47.47 |
| Consensus MFE | -21.54 |
| Energy contribution | -24.48 |
| Covariance contribution | 2.95 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.44 |
| Structure conservation index | 0.45 |
| SVM decision value | 1.00 |
| SVM RNA-class probability | 0.898085 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 1773888 113 - 22224390 AGACCAAUGGACUGGCCCAACCGCUUGUUCGCCAGCGCUGCAGCCUCAGCCUGGUUAGCUUCCUCCUCGGCGGCUGCUACUGCAGCCACCGCUGCAGCGGUACCCUUU---CCAAU .......((((..((....((((((.(....).)))((((((((....(((.((..........))..)))((((((....))))))...)))))))))))..))..)---))).. ( -48.00) >DroSim_CAF1 4628 113 - 1 AGGCCAAUGGACUGGCCCAACCGCUUGUUCGCCAACGCUGCAGCCUCUGCCUGGUUAGCUUCCUCCUCGGCGGCCGCUACUGCAGCCACCGCUGCAGCGCUACCCUUU---CCAAU .(((....((((.(((......))).)))))))..(((((((((....(((.((..........))..)))(((.((....)).)))...))))))))).........---..... ( -41.40) >DroEre_CAF1 6930 113 - 1 AGGCCAAUGGACUGACCCAAUCGCUUGUUCGCCAGCGCUGCAGCCUCGGCUUGGCUGGCUUCCUCCUUGGCGGCCGCUGCAGCCGCCGCCGCUGCAGCGCCACCCUUU---CCAAC .(((.((..(..(((.....))).)..)).))).((((((((((..((((..((((((((.((.....)).)))).....))))))))..))))))))))........---..... ( -47.50) >DroYak_CAF1 7074 113 - 1 AGGCCAAUGGACUGGCCCAAUCGCUUGUUCGCCAGCGCAGCAGCCUCCGCUUGGCUGGCUUCCUCCUCGGCAGCCGCUGCUGCAGCCGCCGCUGCAGCGGCACCCUUG---CCAAU .((((((((((..(((.....((((.(....).)))).....))))))).))))))((((.((.....)).))))(((((((((((....))))))))))).......---..... ( -52.60) >DroMoj_CAF1 12034 113 - 1 AGCCCAAUGCUUUGACCCAAACGCUUAUUGGCCACCACCUGCGCCUCGGCCUUCCGCGCUGCAUCCUCAGCGGCAGCCGCAGCUGCCGCCGCCGCAGC---GCCCUUUGUCCCGCU .(.(((((((((((...)))).))..))))).).......(((....(((.....(((((((.....(.((((((((....)))))))).)..)))))---)).....))).))). ( -38.90) >DroAna_CAF1 7172 110 - 1 AGUCCAAUCGACUGGCCCAGCCGCUUGUUGGCCAGCGCCUGGGCCUCCGCCUGGGUGGCGGCCUCCUCCGCGGCAGCCAUAGCCGCGGCAGCAGCUGCUGUUCCCUUG---CC--- ((((.....))))(((((((.((((.(....).)))).)))))))...((..(((.((((((...(((((((((.......))))))).))..))))))...)))..)---).--- ( -56.40) >consensus AGGCCAAUGGACUGGCCCAACCGCUUGUUCGCCAGCGCUGCAGCCUCCGCCUGGCUGGCUUCCUCCUCGGCGGCCGCUACAGCAGCCGCCGCUGCAGCGGUACCCUUU___CCAAU .......(((((.(((......))).)))))....(((.(((((...(((.......)))........((((((.((....)).))))))))))).)))................. (-21.54 = -24.48 + 2.95)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:49:58 2006