| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 16,002,836 – 16,002,956 |
| Length | 120 |
| Max. P | 0.899724 |

| Location | 16,002,836 – 16,002,956 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.74 |
| Mean single sequence MFE | -26.14 |
| Consensus MFE | -24.26 |
| Energy contribution | -24.14 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.30 |
| Structure conservation index | 0.93 |
| SVM decision value | 0.75 |
| SVM RNA-class probability | 0.839844 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16002836 120 + 22224390 UUAGGAAAAGAAAUCAUUUGCGUUUUAUAUAAACGAGCUACCGAAACGAUAUGUAACGAUACAUUCCAGCGUAGCAGUAGCAAAACUGUAUAUGAAACAGAAUGUCUACAUUUGGUCGCA ..................(((((((((((((.....(((((.(..(((..(((((....))))).....)))..).))))).......)))))))))).(((((....)))))....))) ( -25.90) >DroSec_CAF1 70754 120 + 1 UUAGAAAAUGAAAUCAUUUGCGUUUUAUAUAAACGAGCUAACGAAACGAUUUGUAACGAUACAUUCCAGCGUAGCAGUAGCAAAACUGUGUAUGAAACAGAAUGUCUACAUUUGGUCGCA .....(((((....)))))((((((((((((.....((((..(..(((...((((....))))......)))..)..)))).......)))))))))).(((((....)))))....)). ( -23.60) >DroSim_CAF1 65380 120 + 1 UUAGAAAACGAAAUCAUUUGCGUUUUAUAUAAACGAGCUACCGAAACGAUUUGUAACGAUACAUUCCAGCGUAGCAGUAGCAAAACUGUGUAUGAAACAGAAUGUCUACAUUUGGUCGCA ..................(((((((((((((.....(((((.(..(((...((((....))))......)))..).))))).......)))))))))).(((((....)))))....))) ( -24.60) >DroEre_CAF1 69912 118 + 1 UUAGAAAAAGAAAUCAUUUGCGUUUCAUAUAAACGAGCUACCGAAACGAUUUGUAACGAUACAUUCCUGCGUAGCAGUAGCA--ACUGUGUAUGAAACCGAAUGUCUACAUUUGGUCGCA ..................((((.((((((((.....(((((.(..(((...((((....))))......)))..).))))).--....))))))))((((((((....)))))))))))) ( -30.80) >DroYak_CAF1 53980 118 + 1 UUAGGAAAAGAAAUCAUUUGCGUUUUAUAUAAACGAGCUACCGAAACGAUUUGUAACGAUACAUUCCUGCGUAGCAGUAGCA--ACUGUGUAUGAAACAGAAUGUCUACAUUUGGUCGCA ..................(((((((((((((.....(((((.(..(((...((((....))))......)))..).))))).--....)))))))))).(((((....)))))....))) ( -25.80) >consensus UUAGAAAAAGAAAUCAUUUGCGUUUUAUAUAAACGAGCUACCGAAACGAUUUGUAACGAUACAUUCCAGCGUAGCAGUAGCAAAACUGUGUAUGAAACAGAAUGUCUACAUUUGGUCGCA ..................(((((((((((((.....(((((.(..(((...((((....))))......)))..).))))).......)))))))))).(((((....)))))....))) (-24.26 = -24.14 + -0.12)



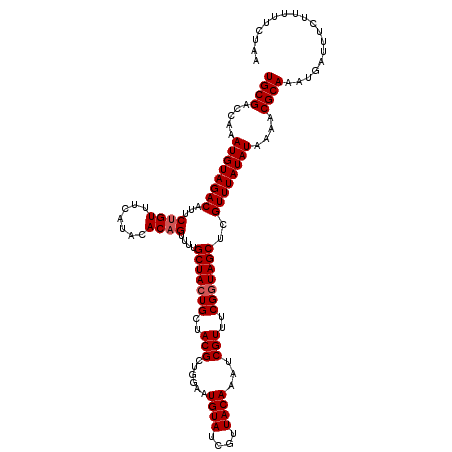
| Location | 16,002,836 – 16,002,956 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.74 |
| Mean single sequence MFE | -25.92 |
| Consensus MFE | -24.16 |
| Energy contribution | -24.56 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.36 |
| Structure conservation index | 0.93 |
| SVM decision value | 1.01 |
| SVM RNA-class probability | 0.899724 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16002836 120 - 22224390 UGCGACCAAAUGUAGACAUUCUGUUUCAUAUACAGUUUUGCUACUGCUACGCUGGAAUGUAUCGUUACAUAUCGUUUCGGUAGCUCGUUUAUAUAAAACGCAAAUGAUUUCUUUUCCUAA ((((.....((((((((...((((.......))))....(((((((..(((.....(((((....)))))..)))..)))))))..))))))))....)))).................. ( -27.90) >DroSec_CAF1 70754 120 - 1 UGCGACCAAAUGUAGACAUUCUGUUUCAUACACAGUUUUGCUACUGCUACGCUGGAAUGUAUCGUUACAAAUCGUUUCGUUAGCUCGUUUAUAUAAAACGCAAAUGAUUUCAUUUUCUAA ........((((...(((((((((.....)))((((...((....))...))))))))))..))))..(((((((((((((...............)))).))))))))).......... ( -21.66) >DroSim_CAF1 65380 120 - 1 UGCGACCAAAUGUAGACAUUCUGUUUCAUACACAGUUUUGCUACUGCUACGCUGGAAUGUAUCGUUACAAAUCGUUUCGGUAGCUCGUUUAUAUAAAACGCAAAUGAUUUCGUUUUCUAA .(((.....((((((((...((((.......))))....(((((((..(((......((((....))))...)))..)))))))..))))))))....)))(((((....)))))..... ( -26.20) >DroEre_CAF1 69912 118 - 1 UGCGACCAAAUGUAGACAUUCGGUUUCAUACACAGU--UGCUACUGCUACGCAGGAAUGUAUCGUUACAAAUCGUUUCGGUAGCUCGUUUAUAUGAAACGCAAAUGAUUUCUUUUUCUAA (((((((.((((....)))).)))((((((......--.(((((((..(((......((((....))))...)))..))))))).......)))))).)))).................. ( -27.94) >DroYak_CAF1 53980 118 - 1 UGCGACCAAAUGUAGACAUUCUGUUUCAUACACAGU--UGCUACUGCUACGCAGGAAUGUAUCGUUACAAAUCGUUUCGGUAGCUCGUUUAUAUAAAACGCAAAUGAUUUCUUUUCCUAA ((((.....((((((((...((((.......)))).--.(((((((..(((......((((....))))...)))..)))))))..))))))))....)))).................. ( -25.90) >consensus UGCGACCAAAUGUAGACAUUCUGUUUCAUACACAGUUUUGCUACUGCUACGCUGGAAUGUAUCGUUACAAAUCGUUUCGGUAGCUCGUUUAUAUAAAACGCAAAUGAUUUCUUUUUCUAA ((((.....((((((((...((((.......))))....(((((((..(((......((((....))))...)))..)))))))..))))))))....)))).................. (-24.16 = -24.56 + 0.40)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:11:28 2006