| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 1,764,960 – 1,765,065 |
| Length | 105 |
| Max. P | 0.962857 |

| Location | 1,764,960 – 1,765,065 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 67.26 |
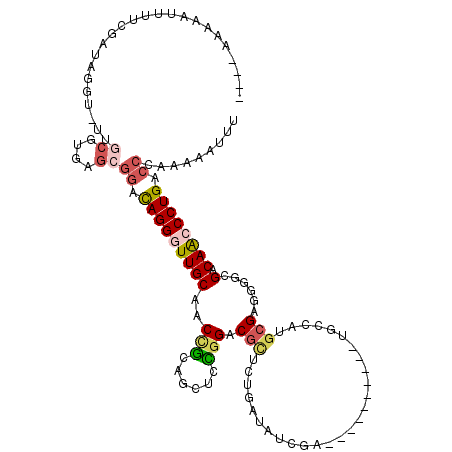
| Mean single sequence MFE | -35.90 |
| Consensus MFE | -13.50 |
| Energy contribution | -14.00 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.31 |
| Mean z-score | -2.24 |
| Structure conservation index | 0.38 |
| SVM decision value | 1.55 |
| SVM RNA-class probability | 0.962857 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
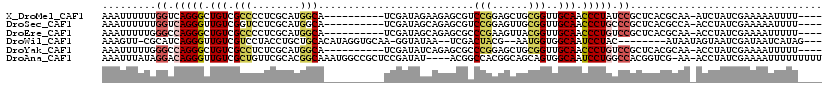
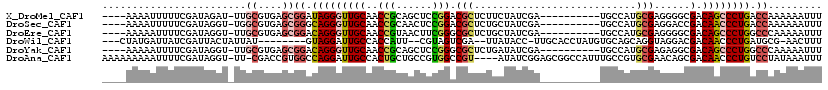
>X_DroMel_CAF1 1764960 105 + 22224390 AAAUUUUUUGGUCAGGGCUGUCGCCCCUCGCAUGGCA----------UCGAUAGAAGAGCGUCCGGAGCUGCGGUUGCAACCCUAUCCGCUCACGCAA-AUCUAUCGAAAAAUUUU---- (((((((...((((((((....))))......)))).----------((((((((...((((.((((...(.(((....))))..))))...))))..-.))))))))))))))).---- ( -32.40) >DroSec_CAF1 6440 105 + 1 AAAUUUUUUGGUCAGGGUUGUCGGUCCUCGCAUGGCA----------UCGAUAGCAGAGCGUCCGGAGUUGCGGUUGCAACCCUGCCCGCUCACGCCA-ACCUAUCGAAAAAUUUU---- ((((((((((((.((((((((((((((......)).)----------)))))))).(((((..(((.(((((....))))).)))..)))))......-.))))))))))))))).---- ( -44.70) >DroEre_CAF1 6098 105 + 1 AAAUUUUUGGGCCAGGGCUGUCGCCCCUCGCAUGGCA----------UCGAUAGCAGAGCGCCCGAAGUUACGGUUGCAACCCUGUCCGCUCACGCAA-ACCUAUCGAAAAUUUUU---- (((((((((((.(((((((((((..((......))..----------.)))))))...(((.(((......))).)))...)))).))((....))..-......)))))))))..---- ( -34.00) >DroWil_CAF1 8525 103 + 1 AAAGUU-CGCAUCAGGGUUGUCGUCCUACCUGCUGCACAUAGGUGCAA-GGUAUAA--UCGACUACG--AAUGGUGGCAAUCCUAC--------AUAAUAGUAAUCGAUAAUCAUAG--- ......-...(((.(((((((((.(((((((..(((((....))))))-))))...--(((....))--)..)))))))))))(((--------......)))...)))........--- ( -27.80) >DroYak_CAF1 6151 105 + 1 AAAUUUUUGGGCCAGGGCUGUCGCCUCUCGCAUGGCA----------UCGAUAUCAGAGCGCCCGGAGCUGCGGUUGCAACCCUGUCCGCUCACGCAA-ACCUAUCGAAAAUUUUU---- ((((((((((...(((..(((.(((........))).----------.........(((((..(((.(.(((....))).).)))..)))))..))).-.))).))))))))))..---- ( -32.10) >DroAna_CAF1 6238 114 + 1 AAAUUUAUAGGACAGGGUUGUCGCUGUUCGCACGGCAAAUGGCCGCUCCGAUAU----ACGGCCACGGCAGCAGUGGCAAUCCUGGCCACGGUCG-AA-ACCUAUCGAAAAUUUUUUUUU ((((((...((.((((((((((((((((.((.((.....((((((.........----.)))))))))))))))))))))))))).))....(((-(.-.....))))))))))...... ( -44.40) >consensus AAAUUUUUGGGUCAGGGCUGUCGCCCCUCGCAUGGCA__________UCGAUAGCAGAGCGCCCGGAGCUGCGGUUGCAACCCUGUCCGCUCACGCAA_ACCUAUCGAAAAUUUUU____ .........((.(((((.(((.(((........)))..........................(((......)))..))).))))).))................................ (-13.50 = -14.00 + 0.50)
| Location | 1,764,960 – 1,765,065 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 67.26 |
| Mean single sequence MFE | -34.85 |
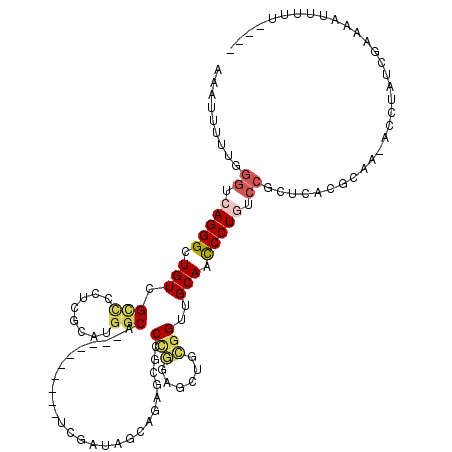
| Consensus MFE | -15.36 |
| Energy contribution | -15.98 |
| Covariance contribution | 0.62 |
| Combinations/Pair | 1.32 |
| Mean z-score | -1.69 |
| Structure conservation index | 0.44 |
| SVM decision value | 0.51 |
| SVM RNA-class probability | 0.764621 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 1764960 105 - 22224390 ----AAAAUUUUUCGAUAGAU-UUGCGUGAGCGGAUAGGGUUGCAACCGCAGCUCCGGACGCUCUUCUAUCGA----------UGCCAUGCGAGGGGCGACAGCCCUGACCAAAAAAUUU ----.(((((((((((((((.-......(((((..(.(((((((....))))))).)..))))).))))))))----------.....((...(((((....)))))...)).))))))) ( -38.70) >DroSec_CAF1 6440 105 - 1 ----AAAAUUUUUCGAUAGGU-UGGCGUGAGCGGGCAGGGUUGCAACCGCAACUCCGGACGCUCUGCUAUCGA----------UGCCAUGCGAGGACCGACAACCCUGACCAAAAAAUUU ----.((((((((.(...(((-(((((.(((((..(.(((((((....))))))).)..))))))))..(((.----------..((......))..)))))))).....).)))))))) ( -34.10) >DroEre_CAF1 6098 105 - 1 ----AAAAAUUUUCGAUAGGU-UUGCGUGAGCGGACAGGGUUGCAACCGUAACUUCGGGCGCUCUGCUAUCGA----------UGCCAUGCGAGGGGCGACAGCCCUGGCCCAAAAAUUU ----........(((((((((-((((....))))))(((((.((..(((......)))))))))).)))))))----------......((..(((((....))))).)).......... ( -39.00) >DroWil_CAF1 8525 103 - 1 ---CUAUGAUUAUCGAUUACUAUUAU--------GUAGGAUUGCCACCAUU--CGUAGUCGA--UUAUACC-UUGCACCUAUGUGCAGCAGGUAGGACGACAACCCUGAUGCG-AACUUU ---.....((.(((((((((....((--------(..((....))..))).--.))))))))--).))...-.(((((....)))))(((..((((........)))).))).-...... ( -25.50) >DroYak_CAF1 6151 105 - 1 ----AAAAAUUUUCGAUAGGU-UUGCGUGAGCGGACAGGGUUGCAACCGCAGCUCCGGGCGCUCUGAUAUCGA----------UGCCAUGCGAGAGGCGACAGCCCUGGCCCAAAAAUUU ----..(((((((........-..((....))((.((((((((....(((..(((((((((.((.......))----------)))).)).)))..))).))))))))..)).))))))) ( -36.60) >DroAna_CAF1 6238 114 - 1 AAAAAAAAAUUUUCGAUAGGU-UU-CGACCGUGGCCAGGAUUGCCACUGCUGCCGUGGCCGU----AUAUCGGAGCGGCCAUUUGCCGUGCGAACAGCGACAACCCUGUCCUAUAAAUUU ......((((((..((((((.-..-.....(((((.......)))))(((((..((((((((----........))))))))((((...)))).))))).....))))))....)))))) ( -35.20) >consensus ____AAAAAUUUUCGAUAGGU_UUGCGUGAGCGGACAGGGUUGCAACCGCAGCUCCGGACGCUCUGAUAUCGA__________UGCCAUGCGAGGGGCGACAACCCUGACCCAAAAAUUU ........................((....))((.(((((((((..(((......))).(((...........................)))......).)))))))).))......... (-15.36 = -15.98 + 0.62)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:49:56 2006