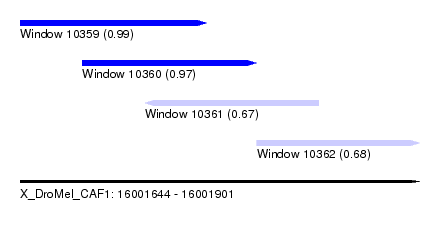
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 16,001,644 – 16,001,901 |
| Length | 257 |
| Max. P | 0.988045 |

| Location | 16,001,644 – 16,001,764 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 97.67 |
| Mean single sequence MFE | -39.58 |
| Consensus MFE | -35.16 |
| Energy contribution | -35.76 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.08 |
| Structure conservation index | 0.89 |
| SVM decision value | 2.10 |
| SVM RNA-class probability | 0.988045 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16001644 120 + 22224390 UGCCCUCGGGGCUCAUAGAAUGAAUCUUAAUGAAUUUCGAUUCGAAAUGUUCAGGAGUCGGCGGUCGACUGCCGGUGGGUGUUACCUUCUGGCAGUAAAGUCAAAAAGUGGCAUCUAAAA ((((((...(((((...((((...((.....))((((((...))))))))))..)))))(((.....((((((((.(((.....))).))))))))...)))....)).))))....... ( -38.60) >DroSec_CAF1 69553 120 + 1 UGCCCUCGGGGCUCAUAGAAUGAAUCUUAAUGAAUUUCGAUUCGAAAUGUUCAGGAGUCGGCGGUCGACUGCCGGUGGGUGUUACCUUCUGGCAGUAAAGUCAAAAAGUGGCAUCUAAAA ((((((...(((((...((((...((.....))((((((...))))))))))..)))))(((.....((((((((.(((.....))).))))))))...)))....)).))))....... ( -38.60) >DroSim_CAF1 63697 120 + 1 UGCCCUCGGGGCUCAUAGAAUGAAUCUUAAUGAAUUUCGAUUCGAAAUGUUCAGGAGUCGGCGGUCGACUGCCGGUGGGUGUUACCUUCUGGCAGUAAAGUCAAAAAGUGGCAUCUAAAA ((((((...(((((...((((...((.....))((((((...))))))))))..)))))(((.....((((((((.(((.....))).))))))))...)))....)).))))....... ( -38.60) >DroEre_CAF1 68683 120 + 1 UGCCUUCGGGGCUCAUAGAAUGAAUCUUAAUGAAUUUCGAUUCGAAAUGUGCAGGAGUCGGCGGUCGACUGCCGGUGGGUGUUACCUUCUGGCAGUAAAGUCAGAAAGUGGCAUCUAAAA ..((....))((.(((.(((((((((.....))..))).))))...))).)).....(((((((....)))))))((((((((((.(((((((......))))))).))))))))))... ( -43.30) >DroYak_CAF1 52748 120 + 1 UGCCUUCGGGGCUCAUAGAAUGAAUCUUAAUGAAUUUCGAUUCGAAAUGUGCAGGAGUCGCCGCUCGACUGCCGGUGGGUGUUACCUUCUGGCAGUAAAGUCCAAAAGUGGCAUCUAAAA ((((...(((((((..(((.....)))...((.((((((...))))))...)).))))).))(((..((((((((.(((.....))).))))))))..)))........))))....... ( -38.80) >consensus UGCCCUCGGGGCUCAUAGAAUGAAUCUUAAUGAAUUUCGAUUCGAAAUGUUCAGGAGUCGGCGGUCGACUGCCGGUGGGUGUUACCUUCUGGCAGUAAAGUCAAAAAGUGGCAUCUAAAA ((((.....(((((...((((...((.....))((((((...))))))))))..)))))(((.....((((((((.(((.....))).))))))))...))).......))))....... (-35.16 = -35.76 + 0.60)



| Location | 16,001,684 – 16,001,796 |
|---|---|
| Length | 112 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.01 |
| Mean single sequence MFE | -35.00 |
| Consensus MFE | -29.77 |
| Energy contribution | -30.27 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.48 |
| Structure conservation index | 0.85 |
| SVM decision value | 1.68 |
| SVM RNA-class probability | 0.971448 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16001684 112 + 22224390 UUCGAAAUGUUCAGGAGUCGGCGGUCGACUGCCGGUGGGUGUUACCUUCUGGCAGUAAAGUCAAAAAGUGGCAUCUAAAAAAAAAAACCCAGACACAUAUGAGUAUAUGAAU-------- .......((((..((..(((((((....)))))))((((((((((.((.((((......)))).)).))))))))))..........))..))))(((((....)))))...-------- ( -32.30) >DroSec_CAF1 69593 110 + 1 UUCGAAAUGUUCAGGAGUCGGCGGUCGACUGCCGGUGGGUGUUACCUUCUGGCAGUAAAGUCAAAAAGUGGCAUCUAAAAA--AACCCCCAGACACAUAUGAGUAUAUGAAU-------- ((((..(((((((.(.(((((.(((..((((((((.(((.....))).))))))))...((((.....)))).........--.))).)).))).)...))))))).)))).-------- ( -32.50) >DroSim_CAF1 63737 110 + 1 UUCGAAAUGUUCAGGAGUCGGCGGUCGACUGCCGGUGGGUGUUACCUUCUGGCAGUAAAGUCAAAAAGUGGCAUCUAAAAA--AAACCCCAGACACAUAUGAGUAUAUGAAU-------- ((((..(((((((.(.(((((.(((..((((((((.(((.....))).))))))))...((((.....)))).........--..))))).))).)...))))))).)))).-------- ( -32.50) >DroEre_CAF1 68723 113 + 1 UUCGAAAUGUGCAGGAGUCGGCGGUCGACUGCCGGUGGGUGUUACCUUCUGGCAGUAAAGUCAGAAAGUGGCAUCUAAAA-----AGCCCAGACACAUAUGACUAUAU--AUGUACGCAC ........((((.((.((((((((....)))))).((((((((((.(((((((......))))))).))))))))))...-----.))))....(((((((....)))--))))..)))) ( -42.70) >consensus UUCGAAAUGUUCAGGAGUCGGCGGUCGACUGCCGGUGGGUGUUACCUUCUGGCAGUAAAGUCAAAAAGUGGCAUCUAAAAA__AAACCCCAGACACAUAUGAGUAUAUGAAU________ .......((((..((..(((((((....)))))))((((((((((.((.((((......)))).)).))))))))))..........))..))))(((((....)))))........... (-29.77 = -30.27 + 0.50)


| Location | 16,001,724 – 16,001,836 |
|---|---|
| Length | 112 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 83.18 |
| Mean single sequence MFE | -27.08 |
| Consensus MFE | -14.42 |
| Energy contribution | -14.54 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.65 |
| Structure conservation index | 0.53 |
| SVM decision value | 0.28 |
| SVM RNA-class probability | 0.665189 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16001724 112 - 22224390 CUGUGUCCACCUAUGUGCGCAUAUGUGUAUAUACAUAUAU--------AUUCAUAUACUCAUAUGUGUCUGGGUUUUUUUUUUUAGAUGCCACUUUUUGACUUUACUGCCAGAAGGUAAC ..(((((.(((((.(.(((((((((.((((((........--------....)))))).)))))))))))))))...........))))).((((((((.(......).))))))))... ( -26.50) >DroSec_CAF1 69633 98 - 1 CUGUGUCCACCUAUGUGC--AUAUGU--------AU--AU--------AUUCAUAUACUCAUAUGUGUCUGGGGGUU--UUUUUAGAUGCCACUUUUUGACUUUACUGCCAGAAGGUAAC ........((((....((--(((((.--------.(--((--------(....))))..))))))).(((((.(((.--...(((((........)))))....))).)))))))))... ( -24.50) >DroSim_CAF1 63777 100 - 1 CUGUGUCCACCUAUGUGC--AUAUGU--------AUAUAU--------AUUCAUAUACUCAUAUGUGUCUGGGGUUU--UUUUUAGAUGCCACUUUUUGACUUUACUGCCAGAAGGUAAC ..(((((..((((.(..(--(((((.--------.(((((--------....)))))..))))))..).))))....--......))))).((((((((.(......).))))))))... ( -23.20) >DroEre_CAF1 68763 109 - 1 CUGUGUCCACCAAUGUAC--AUGUGU--AUAUACACAUAUGUGCGUACAU--AUAUAGUCAUAUGUGUCUGGGCU-----UUUUAGAUGCCACUUUCUGACUUUACUGCCAGAAGGUAAC (((.(((((...((((((--((((((--......))))))))))))((((--((((....)))))))).))))).-----...))).....((((((((.(......).))))))))... ( -34.10) >consensus CUGUGUCCACCUAUGUGC__AUAUGU________AUAUAU________AUUCAUAUACUCAUAUGUGUCUGGGGUUU__UUUUUAGAUGCCACUUUUUGACUUUACUGCCAGAAGGUAAC ............(((((...(((((..........................)))))...)))))(((((((((........))))))))).((((((((.(......).))))))))... (-14.42 = -14.54 + 0.12)

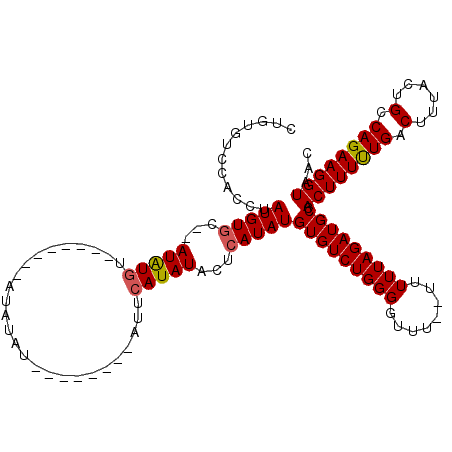

| Location | 16,001,796 – 16,001,901 |
|---|---|
| Length | 105 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 82.65 |
| Mean single sequence MFE | -21.00 |
| Consensus MFE | -18.96 |
| Energy contribution | -19.02 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.41 |
| Structure conservation index | 0.90 |
| SVM decision value | 0.31 |
| SVM RNA-class probability | 0.683180 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16001796 105 + 22224390 AUAUAUGUAUAUACACAUAUGCGCACAUAGGUGGACACAGUUUU----UAAGCGCUUUUCUCUGGCUCUUCCAUCUCGCCAUCAUCGUCCAAUCUGUGGGCCUUUUGUU----------- .(((((((......))))))).((.(((((((((((...(((..----..))).........((((...........)))).....)))).))))))).))........----------- ( -24.70) >DroSec_CAF1 69703 93 + 1 AU--AU--------ACAUAU--GCACAUAGGUGGACACAGUUUU----UAAGCGCUUUUCUCUGGCUCCUCCAUCUCCCCAUCAUCGUCCAAUCUGUGGGCCUUUUGUU----------- ..--..--------(((...--((.(((((((((((...(((..----..)))(((.......)))....................)))).))))))).))....))).----------- ( -19.10) >DroSim_CAF1 63847 106 + 1 AUAUAU--------ACAUAU--GCACAUAGGUGGACACAGUUUU----UAAGCGCUUUUCUCUGGCUCUACCAUCUCCCCAUCAUCGUCCAAUCUGUGGGCCUUUUGUUGUCCCUNNNNN ......--------(((...--((.(((((((((((...(((..----..)))(((.......)))....................)))).))))))).))....)))............ ( -19.10) >DroEre_CAF1 68836 104 + 1 AUAUGUGUAUAU--ACACAU--GUACAUUGGUGGACACAGUUUUUAUUUAAGCGCUUUUCUCUGGCUCUUUCAUCUCCCCAUCAUCGUCCAAUCUGUG-GCCUUUUGUU----------- ((((((((....--))))))--)).(((.(((((((...((((......))))(((.......)))....................)))).))).)))-..........----------- ( -21.10) >consensus AUAUAU________ACAUAU__GCACAUAGGUGGACACAGUUUU____UAAGCGCUUUUCUCUGGCUCUUCCAUCUCCCCAUCAUCGUCCAAUCUGUGGGCCUUUUGUU___________ ..............(((.....((.(((((((((((...((((......))))(((.......)))....................)))).))))))).))....)))............ (-18.96 = -19.02 + 0.06)

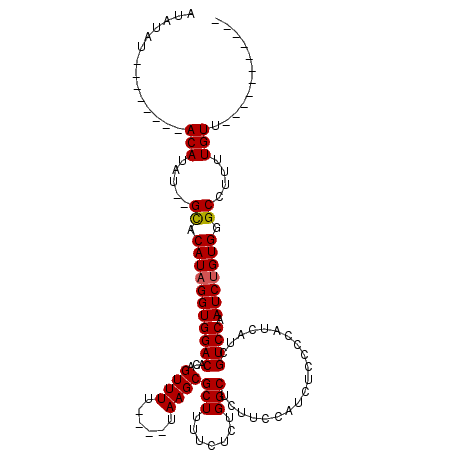
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:11:26 2006