
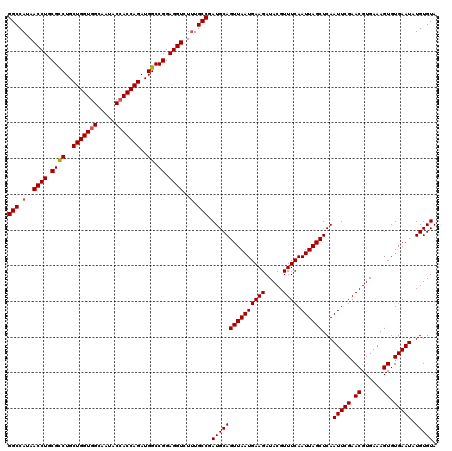
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 15,998,662 – 15,998,822 |
| Length | 160 |
| Max. P | 0.980959 |

| Location | 15,998,662 – 15,998,782 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 98.67 |
| Mean single sequence MFE | -38.26 |
| Consensus MFE | -37.50 |
| Energy contribution | -37.74 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.02 |
| Mean z-score | -1.68 |
| Structure conservation index | 0.98 |
| SVM decision value | 1.80 |
| SVM RNA-class probability | 0.977847 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15998662 120 - 22224390 GGCCAUAACCUGCGUCUGCUGGUGGCAAUACCACCAGAUGGCCGGAGGUCUUUGCCGAUGCAGUUAAUGAAGAUACGUUUCAAUUAGCUCAAUUCGAACGUGAAAGUGUGAAUAUGUGUA (((.(..((((.((.((((((((((.....))))))).))).)).))))..).))).(((((((((((((((.....)))).))))))...(((((.((......)).))))).))))). ( -38.40) >DroSec_CAF1 66671 120 - 1 GGCCAUAACCUGCGCCUGCUGGUGGCAAUACCACCAGAUGGCCGGAGGUCUUUGCCGAUGCAGUUAAUGAAGAUACGUUUCAAUUAGCUCAAUUCGAACGUGAAAGUGUGAAUAUGUGUA (((.(..((((.((((..(((((((.....)))))))..)).)).))))..).))).(((((((((((((((.....)))).))))))...(((((.((......)).))))).))))). ( -39.80) >DroSim_CAF1 60838 120 - 1 GGCCAUAACCUGCGCCUGCUGGUGGCAAUACCACCAGAUGGCCGGAGGUCUUUGCCGAUGCAGUUAAUGAAGAUACGUUUCAAUUAGCUCAAUUCGAACGUGAAAGUGUGAAUAUGUGUA (((.(..((((.((((..(((((((.....)))))))..)).)).))))..).))).(((((((((((((((.....)))).))))))...(((((.((......)).))))).))))). ( -39.80) >DroEre_CAF1 65684 120 - 1 GGCCAUAACCUGCGCCUGCUGGUGGCAAUACCACCAGAUGGCCGGAGGUCUCUGCCGAUGCAGUUAAUGAAGAUACGUUUCAAUUAGCUCAAUUCGAACGUGAAAGUGUGAAUAUGUGUA (((....((((.((((..(((((((.....)))))))..)).)).))))....))).(((((((((((((((.....)))).))))))...(((((.((......)).))))).))))). ( -39.10) >DroYak_CAF1 49875 120 - 1 GGCCAUAACCUGCGCCUGCUGGUGGCACUACAACCAGAUGGCCGGAGGUCUUUGCCGAUGCAGUUAAUGAAGAUACGUUUCAAUUAGCUCAAUUCGAACGUGAAAGUGUGAAUAUGUGUA (((.(..((((.((((..((((((......).)))))..)).)).))))..).))).(((((((((((((((.....)))).))))))...(((((.((......)).))))).))))). ( -34.20) >consensus GGCCAUAACCUGCGCCUGCUGGUGGCAAUACCACCAGAUGGCCGGAGGUCUUUGCCGAUGCAGUUAAUGAAGAUACGUUUCAAUUAGCUCAAUUCGAACGUGAAAGUGUGAAUAUGUGUA (((.(..((((.((((..(((((((.....)))))))..)).)).))))..).))).(((((((((((((((.....)))).))))))...(((((.((......)).))))).))))). (-37.50 = -37.74 + 0.24)


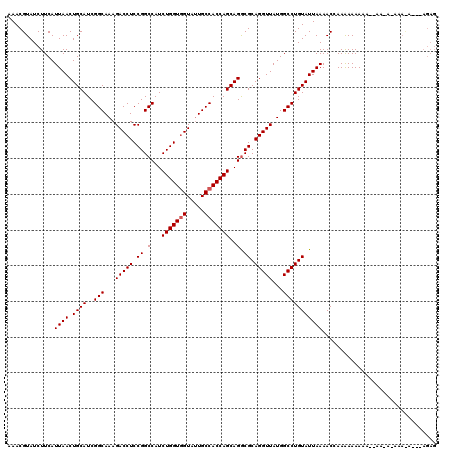
| Location | 15,998,702 – 15,998,822 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 84.83 |
| Mean single sequence MFE | -29.87 |
| Consensus MFE | -27.13 |
| Energy contribution | -27.47 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.49 |
| Structure conservation index | 0.91 |
| SVM decision value | 1.88 |
| SVM RNA-class probability | 0.980959 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15998702 120 + 22224390 AAACGUAUCUUCAUUAACUGCAUCGGCAAAGACCUCCGGCCAUCUGGUGGUAUUGCCACCAGCAGACGCAGGUUAUGGCCUGUAUUAAAACCAAGAAAAAAAAAAAAAAAACACACAGAG .......((((......((((...(((((..(((.((((....)))).))).)))))....))))..((((((....)))))).........))))........................ ( -29.90) >DroSim_CAF1 60878 109 + 1 AAACGUAUCUUCAUUAACUGCAUCGGCAAAGACCUCCGGCCAUCUGGUGGUAUUGCCACCAGCAGGCGCAGGUUAUGGCCUGUAUUAAAACCAAAAAAAAAGGAA-----------AGAG .......((((.......(((....)))......(((.(((..(((((((.....)))))))..)))((((((....))))))..................))))-----------))). ( -32.70) >DroYak_CAF1 49915 108 + 1 AAACGUAUCUUCAUUAACUGCAUCGGCAAAGACCUCCGGCCAUCUGGUUGUAGUGCCACCAGCAGGCGCAGGUUAUGGCCUGUAUUAAAACAAAAAAAAG----A-AUAAAUA------- .......((((......((((...((((...((..((((....))))..))..))))....))))..((((((....))))))..............)))----)-.......------- ( -27.00) >consensus AAACGUAUCUUCAUUAACUGCAUCGGCAAAGACCUCCGGCCAUCUGGUGGUAUUGCCACCAGCAGGCGCAGGUUAUGGCCUGUAUUAAAACCAAAAAAAAA__AA_A_AAA_A___AGAG .............((((.((((..(((...(((((.((.(...(((((((.....)))))))..).)).)))))...)))))))))))................................ (-27.13 = -27.47 + 0.33)


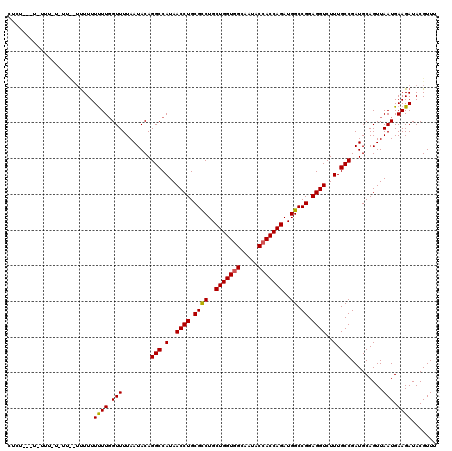
| Location | 15,998,702 – 15,998,822 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 84.83 |
| Mean single sequence MFE | -31.23 |
| Consensus MFE | -28.40 |
| Energy contribution | -28.30 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.56 |
| Structure conservation index | 0.91 |
| SVM decision value | 0.76 |
| SVM RNA-class probability | 0.842327 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15998702 120 - 22224390 CUCUGUGUGUUUUUUUUUUUUUUUUCUUGGUUUUAAUACAGGCCAUAACCUGCGUCUGCUGGUGGCAAUACCACCAGAUGGCCGGAGGUCUUUGCCGAUGCAGUUAAUGAAGAUACGUUU ......((((((((.............((((((......))))))((((.((((((..(((((((.....)))))))..(((.((....))..)))))))))))))..)))))))).... ( -36.40) >DroSim_CAF1 60878 109 - 1 CUCU-----------UUCCUUUUUUUUUGGUUUUAAUACAGGCCAUAACCUGCGCCUGCUGGUGGCAAUACCACCAGAUGGCCGGAGGUCUUUGCCGAUGCAGUUAAUGAAGAUACGUUU ....-----------...((((....(((((......)).(((.(..((((.((((..(((((((.....)))))))..)).)).))))..).)))....))).....))))........ ( -31.10) >DroYak_CAF1 49915 108 - 1 -------UAUUUAU-U----CUUUUUUUUGUUUUAAUACAGGCCAUAACCUGCGCCUGCUGGUGGCACUACAACCAGAUGGCCGGAGGUCUUUGCCGAUGCAGUUAAUGAAGAUACGUUU -------.......-(----((((...((((.........(((.(..((((.((((..((((((......).)))))..)).)).))))..).)))...)))).....)))))....... ( -26.20) >consensus CUCU___U_UUU_U_UU__UUUUUUUUUGGUUUUAAUACAGGCCAUAACCUGCGCCUGCUGGUGGCAAUACCACCAGAUGGCCGGAGGUCUUUGCCGAUGCAGUUAAUGAAGAUACGUUU ........................((((.(((........(((.(..((((.((((..(((((((.....)))))))..)).)).))))..).))).........))).))))....... (-28.40 = -28.30 + -0.11)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:11:17 2006