
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 15,993,075 – 15,993,329 |
| Length | 254 |
| Max. P | 0.992512 |

| Location | 15,993,075 – 15,993,185 |
|---|---|
| Length | 110 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 88.63 |
| Mean single sequence MFE | -33.88 |
| Consensus MFE | -30.03 |
| Energy contribution | -29.91 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.20 |
| Structure conservation index | 0.89 |
| SVM decision value | 1.65 |
| SVM RNA-class probability | 0.970006 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15993075 110 + 22224390 U-------UCCAGAUGCGAGUAAUAACGUCGCAGUCAUGAAUUGAUAAUCGGAUGGCAAUGCAAGUG--GUGGGCUCAAGGGCAUUGCAAAA-AGGCGGGAAAGGUCACCGCCAAAUAAU .-------(((...(((((((....)).)))))((((.....))))....)))..(((((((...((--(.....)))...)))))))....-.(((((((....)).)))))....... ( -35.00) >DroSec_CAF1 61001 112 + 1 U-------UCCAGAUGCGAGUAAUAACGUCGCAGUCAUGAAUUGAUAAUCGGUUGGCAAUGCAAGUGGGGUGGGCUCAAGGGCAUUGCAAAA-AGGCGGGAAAGGUCACCGCCAAAUAAU .-------.((...(((((((....)).)))))((((.....))))....))...(((((((...((((.....))))...)))))))....-.(((((((....)).)))))....... ( -36.20) >DroEre_CAF1 59689 112 + 1 UUCCAUUUUCCAGAUGCGAGUAAUAACGUCGCAGUCAUGAAUUGAUAAUCGGAUGGCAGUGCAAGA-----GGGCUCAAGGGCAUUGCGAAAAGGGCGGGAAAGGUCACCGCCGAAU--- (((.....(((...(((((((....)).)))))((((.....))))....)))..(((((((..((-----....))....))))))))))...(((((((....)).)))))....--- ( -33.20) >DroYak_CAF1 44043 105 + 1 U-------UCCAGAUGCGAGUAAUAACGUCGCAGUCAUGAAUUGAUAAUCGGAUGGCAAUGCAAGA-----GGGCUCAAGGGCAUUGCAAAAAAGGCGGGAAAGGUCACUGCCGAAU--- .-------(((...(((((((....)).)))))((((.....))))....)))..(((((((..((-----....))....)))))))......(((((((....)).)))))....--- ( -31.10) >consensus U_______UCCAGAUGCGAGUAAUAACGUCGCAGUCAUGAAUUGAUAAUCGGAUGGCAAUGCAAGA_____GGGCUCAAGGGCAUUGCAAAA_AGGCGGGAAAGGUCACCGCCAAAU___ ........(((...(((((((....)).)))))((((.....))))....)))..(((((((...................)))))))......(((((((....)).)))))....... (-30.03 = -29.91 + -0.12)



| Location | 15,993,075 – 15,993,185 |
|---|---|
| Length | 110 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 88.63 |
| Mean single sequence MFE | -27.15 |
| Consensus MFE | -24.20 |
| Energy contribution | -24.70 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.83 |
| Structure conservation index | 0.89 |
| SVM decision value | 1.02 |
| SVM RNA-class probability | 0.902264 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15993075 110 - 22224390 AUUAUUUGGCGGUGACCUUUCCCGCCU-UUUUGCAAUGCCCUUGAGCCCAC--CACUUGCAUUGCCAUCCGAUUAUCAAUUCAUGACUGCGACGUUAUUACUCGCAUCUGGA-------A .......(((((.((....))))))).-....(((((((...((.......--))...)))))))..(((((........))..((.(((((.((....))))))))).)))-------. ( -29.10) >DroSec_CAF1 61001 112 - 1 AUUAUUUGGCGGUGACCUUUCCCGCCU-UUUUGCAAUGCCCUUGAGCCCACCCCACUUGCAUUGCCAACCGAUUAUCAAUUCAUGACUGCGACGUUAUUACUCGCAUCUGGA-------A .......(((((.((....))))))).-....(((((((...((.........))...)))))))..............((((.((.(((((.((....)))))))))))))-------. ( -27.90) >DroEre_CAF1 59689 112 - 1 ---AUUCGGCGGUGACCUUUCCCGCCCUUUUCGCAAUGCCCUUGAGCCC-----UCUUGCACUGCCAUCCGAUUAUCAAUUCAUGACUGCGACGUUAUUACUCGCAUCUGGAAAAUGGAA ---....(((((.((....)))))))......(((.(((....((....-----))..))).)))..((((.....)..((((.((.(((((.((....)))))))))))))....))). ( -27.50) >DroYak_CAF1 44043 105 - 1 ---AUUCGGCAGUGACCUUUCCCGCCUUUUUUGCAAUGCCCUUGAGCCC-----UCUUGCAUUGCCAUCCGAUUAUCAAUUCAUGACUGCGACGUUAUUACUCGCAUCUGGA-------A ---..((((..(((........))).......(((((((....((....-----))..)))))))...)))).......((((.((.(((((.((....)))))))))))))-------. ( -24.10) >consensus ___AUUCGGCGGUGACCUUUCCCGCCU_UUUUGCAAUGCCCUUGAGCCC_____ACUUGCAUUGCCAUCCGAUUAUCAAUUCAUGACUGCGACGUUAUUACUCGCAUCUGGA_______A .......(((((.((....)))))))......(((((((....(((.........))))))))))..............((((.((.(((((.((....)))))))))))))........ (-24.20 = -24.70 + 0.50)



| Location | 15,993,108 – 15,993,220 |
|---|---|
| Length | 112 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 87.86 |
| Mean single sequence MFE | -34.25 |
| Consensus MFE | -30.32 |
| Energy contribution | -29.76 |
| Covariance contribution | -0.56 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.69 |
| Structure conservation index | 0.89 |
| SVM decision value | 2.33 |
| SVM RNA-class probability | 0.992512 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15993108 112 + 22224390 AUUGAUAAUCGGAUGGCAAUGCAAGUG--GUGGGCUCAAGGGCAUUGCAAAA-AGGCGGGAAAGGUCACCGCCAAAUAAUGGUGG--ACCCAUCA---GAGGAGGGUAAUGUGUCAAAUG .((((((((((....(((((((...((--(.....)))...)))))))....-.(((((((....)).)))))......))))..--((((.((.---...))))))....))))))... ( -34.40) >DroSec_CAF1 61034 116 + 1 AUUGAUAAUCGGUUGGCAAUGCAAGUGGGGUGGGCUCAAGGGCAUUGCAAAA-AGGCGGGAAAGGUCACCGCCAAAUAAUGAUGGAGACCCAUCA---GAGGAGGGUAAUGUGUCAAAUG .((((((.((.(((.(((((((...((((.....))))...)))))))....-.(((((((....)).))))).......))).)).((((.((.---...))))))....))))))... ( -39.30) >DroEre_CAF1 59729 107 + 1 AUUGAUAAUCGGAUGGCAGUGCAAGA-----GGGCUCAAGGGCAUUGCGAAAAGGGCGGGAAAGGUCACCGCCGAAU---GGUGG--ACCCAUCG---UUGGAGGGUAAUGUGUCAAAUG .((((((((((....(((((((..((-----....))....)))))))......(((((((....)).)))))...)---)))..--((((.((.---...))))))....))))))... ( -32.70) >DroYak_CAF1 44076 110 + 1 AUUGAUAAUCGGAUGGCAAUGCAAGA-----GGGCUCAAGGGCAUUGCAAAAAAGGCGGGAAAGGUCACUGCCGAAU---GGUGG--ACCCAUCAGAGGAGGAGGGUAAUGUGUCAAAUG .((((((((((....(((((((..((-----....))....)))))))......(((((((....)).)))))...)---)))..--((((..(......)..))))....))))))... ( -30.60) >consensus AUUGAUAAUCGGAUGGCAAUGCAAGA_____GGGCUCAAGGGCAUUGCAAAA_AGGCGGGAAAGGUCACCGCCAAAU___GGUGG__ACCCAUCA___GAGGAGGGUAAUGUGUCAAAUG .(((((((((.....(((((((...................)))))))......(((((((....)).))))).......)))....((((..(......)..))))....))))))... (-30.32 = -29.76 + -0.56)



| Location | 15,993,108 – 15,993,220 |
|---|---|
| Length | 112 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 87.86 |
| Mean single sequence MFE | -23.75 |
| Consensus MFE | -21.00 |
| Energy contribution | -21.50 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.19 |
| Structure conservation index | 0.88 |
| SVM decision value | 1.62 |
| SVM RNA-class probability | 0.968285 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15993108 112 - 22224390 CAUUUGACACAUUACCCUCCUC---UGAUGGGU--CCACCAUUAUUUGGCGGUGACCUUUCCCGCCU-UUUUGCAAUGCCCUUGAGCCCAC--CACUUGCAUUGCCAUCCGAUUAUCAAU ...((((...............---((((((..--...))))))...(((((.((....))))))).-....(((((((...((.......--))...)))))))..........)))). ( -25.70) >DroSec_CAF1 61034 116 - 1 CAUUUGACACAUUACCCUCCUC---UGAUGGGUCUCCAUCAUUAUUUGGCGGUGACCUUUCCCGCCU-UUUUGCAAUGCCCUUGAGCCCACCCCACUUGCAUUGCCAACCGAUUAUCAAU ...((((...............---((((((....))))))......(((((.((....))))))).-....(((((((...((.........))...)))))))..........)))). ( -26.20) >DroEre_CAF1 59729 107 - 1 CAUUUGACACAUUACCCUCCAA---CGAUGGGU--CCACC---AUUCGGCGGUGACCUUUCCCGCCCUUUUCGCAAUGCCCUUGAGCCC-----UCUUGCACUGCCAUCCGAUUAUCAAU ...((((......((((((...---.)).))))--.....---..(((((((.((....)))))))......(((.(((....((....-----))..))).))).....))...)))). ( -22.20) >DroYak_CAF1 44076 110 - 1 CAUUUGACACAUUACCCUCCUCCUCUGAUGGGU--CCACC---AUUCGGCAGUGACCUUUCCCGCCUUUUUUGCAAUGCCCUUGAGCCC-----UCUUGCAUUGCCAUCCGAUUAUCAAU ...((((......((((..(......)..))))--.....---..((((..(((........))).......(((((((....((....-----))..)))))))...))))...)))). ( -20.90) >consensus CAUUUGACACAUUACCCUCCUC___UGAUGGGU__CCACC___AUUCGGCGGUGACCUUUCCCGCCU_UUUUGCAAUGCCCUUGAGCCC_____ACUUGCAUUGCCAUCCGAUUAUCAAU ...((((......((((..(......)..))))..............(((((.((....)))))))......(((((((....(((.........))))))))))..........)))). (-21.00 = -21.50 + 0.50)


| Location | 15,993,146 – 15,993,259 |
|---|---|
| Length | 113 |
| Sequences | 4 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 87.48 |
| Mean single sequence MFE | -38.65 |
| Consensus MFE | -35.53 |
| Energy contribution | -36.15 |
| Covariance contribution | 0.62 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.76 |
| Structure conservation index | 0.92 |
| SVM decision value | 1.01 |
| SVM RNA-class probability | 0.899931 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15993146 113 + 22224390 GGCAUUGCAAAA-AGGCGGGAAAGGUCACCGCCAAAUAAUGGUGG--ACCCAUCA---GAGGAGGGUAAUGUGUCAAAUGAGCACCCCGGGGUAGCGAUUUUCCCAGGGGACUUCAGGU .(((((((....-.(((((((....)).)))))......(((((.--...)))))---.......)))))))......((((..((((((((.........)))).))))..))))... ( -38.30) >DroSec_CAF1 61074 115 + 1 GGCAUUGCAAAA-AGGCGGGAAAGGUCACCGCCAAAUAAUGAUGGAGACCCAUCA---GAGGAGGGUAAUGUGUCAAAUGAGCAACCUGGGGUAGCGAUUUUCCAAGGGGACUUCAGGC .(((((((....-.(((((((....)).)))))......((((((....))))))---.......))))))).............(((((((.........)))(((....))))))). ( -35.30) >DroEre_CAF1 59764 111 + 1 GGCAUUGCGAAAAGGGCGGGAAAGGUCACCGCCGAAU---GGUGG--ACCCAUCG---UUGGAGGGUAAUGUGUCAAAUGAGCACCCUGGGGUACCGAUUUUCCCAGGGGAUUGCAGGU .(((((((......(((((((....)).))))).(((---((((.--...)))))---)).....))))))).........(((((((((((.........))))))))...))).... ( -42.40) >DroYak_CAF1 44111 114 + 1 GGCAUUGCAAAAAAGGCGGGAAAGGUCACUGCCGAAU---GGUGG--ACCCAUCAGAGGAGGAGGGUAAUGUGUCAAAUGAGCACCCUGGGGUACCGAUUUUCCCAGGGGAUUUCACUG .(((((((......(((((((....)).)))))...(---((((.--...)))))..........)))))))......((((..((((((((.........))))))))...))))... ( -38.60) >consensus GGCAUUGCAAAA_AGGCGGGAAAGGUCACCGCCAAAU___GGUGG__ACCCAUCA___GAGGAGGGUAAUGUGUCAAAUGAGCACCCUGGGGUACCGAUUUUCCCAGGGGACUUCAGGU .(((((((......(((((((....)).))))).......(((((....)))))...........)))))))......((((..((((((((.........))))))))...))))... (-35.53 = -36.15 + 0.62)



| Location | 15,993,146 – 15,993,259 |
|---|---|
| Length | 113 |
| Sequences | 4 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 87.48 |
| Mean single sequence MFE | -30.05 |
| Consensus MFE | -26.04 |
| Energy contribution | -26.60 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.37 |
| Structure conservation index | 0.87 |
| SVM decision value | 0.02 |
| SVM RNA-class probability | 0.545949 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15993146 113 - 22224390 ACCUGAAGUCCCCUGGGAAAAUCGCUACCCCGGGGUGCUCAUUUGACACAUUACCCUCCUC---UGAUGGGU--CCACCAUUAUUUGGCGGUGACCUUUCCCGCCU-UUUUGCAAUGCC ...(((.(..(((((((...........)))))))..)))).......((((.........---((((((..--...))))))...(((((.((....))))))).-......)))).. ( -33.30) >DroSec_CAF1 61074 115 - 1 GCCUGAAGUCCCCUUGGAAAAUCGCUACCCCAGGUUGCUCAUUUGACACAUUACCCUCCUC---UGAUGGGUCUCCAUCAUUAUUUGGCGGUGACCUUUCCCGCCU-UUUUGCAAUGCC ((.(((.((..(((.((...........)).)))..))))).(((.((.............---((((((....))))))......(((((.((....))))))).-...))))).)). ( -25.30) >DroEre_CAF1 59764 111 - 1 ACCUGCAAUCCCCUGGGAAAAUCGGUACCCCAGGGUGCUCAUUUGACACAUUACCCUCCAA---CGAUGGGU--CCACC---AUUCGGCGGUGACCUUUCCCGCCCUUUUCGCAAUGCC ....(((...(((((((...........)))))))(((..............((((((...---.)).))))--.....---....(((((.((....)))))))......))).))). ( -33.00) >DroYak_CAF1 44111 114 - 1 CAGUGAAAUCCCCUGGGAAAAUCGGUACCCCAGGGUGCUCAUUUGACACAUUACCCUCCUCCUCUGAUGGGU--CCACC---AUUCGGCAGUGACCUUUCCCGCCUUUUUUGCAAUGCC ..(..(((.....(((((((((.((((((....)))))).))))..(((...((((..(......)..))))--...((---....))..))).....)))))....)))..)...... ( -28.60) >consensus ACCUGAAAUCCCCUGGGAAAAUCGCUACCCCAGGGUGCUCAUUUGACACAUUACCCUCCUC___UGAUGGGU__CCACC___AUUCGGCGGUGACCUUUCCCGCCU_UUUUGCAAUGCC ..........(((((((...........)))))))(((..............((((..(......)..))))..............(((((.((....)))))))......)))..... (-26.04 = -26.60 + 0.56)

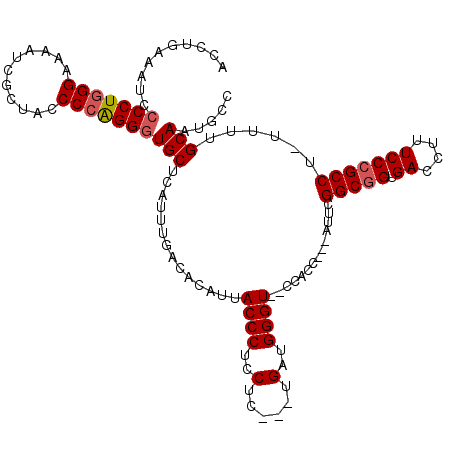

| Location | 15,993,220 – 15,993,329 |
|---|---|
| Length | 109 |
| Sequences | 4 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 72.10 |
| Mean single sequence MFE | -29.93 |
| Consensus MFE | -17.90 |
| Energy contribution | -18.90 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.75 |
| Structure conservation index | 0.60 |
| SVM decision value | 0.86 |
| SVM RNA-class probability | 0.867882 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15993220 109 + 22224390 AGCACCCCGGGGUAGCGAUUUUCCCAGGGGACUUCAGGUUACUGAUCUCAUACAGUCCG--AACUGGUUGAAUUUGCAACUGAAUAUUUUGUAUUAAGAUAAUACUCA--ACA .........((((((((((((..((((.(((((((((....))))........))))).--..))))..))).)))).......(((((((...))))))).))))).--... ( -26.50) >DroSec_CAF1 61150 110 + 1 AGCAACCUGGGGUAGCGAUUUUCCAAGGGGACUUCAGGCUACUGAUCUCAUACAGUCUA-AAACUGGGGGAAUUUGCUACUGAAUAUCUUGUAUUAAGAUGAUACUCA--ACA .......(((((((((((.(((((...((((..((((....))))))))...((((...-..)))).))))).)))))))....(((((((...)))))))...))))--... ( -32.10) >DroEre_CAF1 59836 95 + 1 AGCACCCUGGGGUACCGAUUUUCCCAGGGGAUUGCAGGUUACUGAUCUCCUGCUGCCAC--AGGCCGCGGAAUUUGCUACUGAAUG----------------CACUCAAAACA .(((((((((((.........))))))))....(((((((.(((....((((......)--)))...)))))))))).......))----------------).......... ( -32.30) >DroYak_CAF1 44186 97 + 1 AGCACCCUGGGGUACCGAUUUUCCCAGGGGAUUUCACUGUAGCGAUCUCAUACUGCCGCUCAACUGGUGGCAUUUGCUUCUGAUUA----------------CACUCAAAACA ((((((((((((.........))))))))((..((.(....).))..))....(((((((.....)))))))..))))........----------------........... ( -28.80) >consensus AGCACCCUGGGGUACCGAUUUUCCCAGGGGACUUCAGGUUACUGAUCUCAUACAGCCAC__AACUGGUGGAAUUUGCUACUGAAUA________________CACUCA__ACA .(((((((((((.........))))))))((..((((....))))..))...(((((........)))))....))).................................... (-17.90 = -18.90 + 1.00)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:11:09 2006