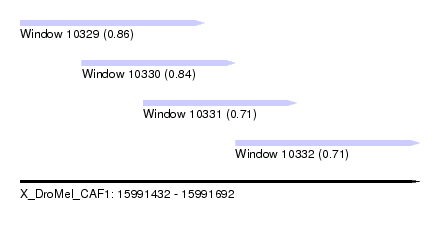
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 15,991,432 – 15,991,692 |
| Length | 260 |
| Max. P | 0.857005 |

| Location | 15,991,432 – 15,991,552 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 97.99 |
| Mean single sequence MFE | -29.38 |
| Consensus MFE | -26.00 |
| Energy contribution | -26.00 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.61 |
| Structure conservation index | 0.88 |
| SVM decision value | 0.81 |
| SVM RNA-class probability | 0.857005 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15991432 120 + 22224390 CAUCAACCGUCAACAUCGCGGCCCUGUCGCAUAAUAUGAUAAUAUGAUAAUAUGGCAAUCGCCCCAGGUAGAGACAACAAAGGGAUCUGUAUCUGUCCGCGACAGAUACAACUUCUACUA ..((.((((((......(((((...)))))...((((....))))))).....(((....)))...))).)).........((((..((((((((((...))))))))))..)))).... ( -30.70) >DroSec_CAF1 59318 119 + 1 CAUCAACCGUCAACGACGCGGCCCUGUCGCAUAAUAUGAUAAUAUGAUAAUAUG-CAAUCGCCCCAGGUAGAGACAACAAAGGGAUCUGUAUCUGUCCGCGACAGAUACUACUUCUACUA ......((((.......))))((((((((((((.(((.((...)).))).))))-)..(((((...))).))))).....))))....(((((((((...)))))))))........... ( -28.60) >DroSim_CAF1 50394 119 + 1 CAUCAACCGUCAACGACGCGGCCCUGUCGCAUAAUAUGAUAAUAUGAUAAUAUG-CAAUCGCCCCAGGUAGAGACAACAAAGGGAUCUGUAUCUGUCCGCGACAGAUACUACUUCUACUA ......((((.......))))((((((((((((.(((.((...)).))).))))-)..(((((...))).))))).....))))....(((((((((...)))))))))........... ( -28.60) >DroEre_CAF1 58210 119 + 1 CAUCAACCGUCGACAUCGCGGCCCUGUCGCAUAAUAUGAUAAUAUGAUAAUAUG-CAAUCGCCCCAGGUAGAGACAACAAAGGGAUCUGUAUCUGUCCGCGACAGAUACUACUUCUACUA ......(((.((....)))))((((((((((((.(((.((...)).))).))))-)..(((((...))).))))).....))))....(((((((((...)))))))))........... ( -30.40) >DroYak_CAF1 42420 119 + 1 CAUCAACCGUCAACAUCGCGGCCCUGUCGCAUAAUAUGAUAAUAUGAUAAUAUG-CAAUCGCCCCAGGUAGAGACAACAAAGGGAUCUGUAUCUGUCCGCGACAGAUACUACUUCUACUA ......((((.......))))((((((((((((.(((.((...)).))).))))-)..(((((...))).))))).....))))....(((((((((...)))))))))........... ( -28.60) >consensus CAUCAACCGUCAACAUCGCGGCCCUGUCGCAUAAUAUGAUAAUAUGAUAAUAUG_CAAUCGCCCCAGGUAGAGACAACAAAGGGAUCUGUAUCUGUCCGCGACAGAUACUACUUCUACUA ......((((.......))))..(((((((.......(((.((((....))))....)))....((((((.((((........).))).))))))...)))))))............... (-26.00 = -26.00 + 0.00)



| Location | 15,991,472 – 15,991,572 |
|---|---|
| Length | 100 |
| Sequences | 4 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 92.75 |
| Mean single sequence MFE | -26.90 |
| Consensus MFE | -21.52 |
| Energy contribution | -21.53 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.22 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.75 |
| SVM RNA-class probability | 0.841928 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15991472 100 + 22224390 AAUAUGAUAAUAUGGCAAUCGCCCCAGGUAGAGACAACAAAGGGAUCUGUAUCUGUCCGCGACAGAUACAACUUCUACUAC--------CAACUCAACGGCGAAGGCG ..............((..(((((...(((((.(....)...((((..((((((((((...))))))))))..)))).))))--------)........)))))..)). ( -29.10) >DroSec_CAF1 59358 99 + 1 AAUAUGAUAAUAUG-CAAUCGCCCCAGGUAGAGACAACAAAGGGAUCUGUAUCUGUCCGCGACAGAUACUACUUCUACUAC--------CAACUCAACGGCGAAGGCG .............(-(..(((((...((((((((......((....))(((((((((...)))))))))....))).))))--------)........)))))..)). ( -27.60) >DroSim_CAF1 50434 99 + 1 AAUAUGAUAAUAUG-CAAUCGCCCCAGGUAGAGACAACAAAGGGAUCUGUAUCUGUCCGCGACAGAUACUACUUCUACUAC--------CAACUCAACGGCGAAGGCG .............(-(..(((((...((((((((......((....))(((((((((...)))))))))....))).))))--------)........)))))..)). ( -27.60) >DroEre_CAF1 58250 104 + 1 AAUAUGAUAAUAUG-CAAUCGCCCCAGGUAGAGACAACAAAGGGAUCUGUAUCUGUCCGCGACAGAUACUACUUCUACUACAGUAAAACCAAGUAAACGGCGAAG--- ..............-...(((((...(((((((.......((....))(((((((((...)))))))))...))))))).............(....))))))..--- ( -23.30) >consensus AAUAUGAUAAUAUG_CAAUCGCCCCAGGUAGAGACAACAAAGGGAUCUGUAUCUGUCCGCGACAGAUACUACUUCUACUAC________CAACUCAACGGCGAAGGCG ..................(((((......(((..(.......)..)))(((((((((...))))))))).............................)))))..... (-21.52 = -21.53 + 0.00)

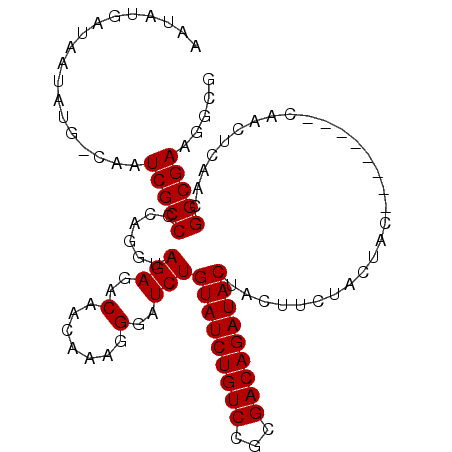

| Location | 15,991,512 – 15,991,612 |
|---|---|
| Length | 100 |
| Sequences | 4 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 88.94 |
| Mean single sequence MFE | -32.62 |
| Consensus MFE | -27.98 |
| Energy contribution | -28.10 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.55 |
| Structure conservation index | 0.86 |
| SVM decision value | 0.36 |
| SVM RNA-class probability | 0.705390 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15991512 100 + 22224390 AGGGAUCUGUAUCUGUCCGCGACAGAUACAACUUCUACUAC--------CAACUCAACGGCGAAGGCGAAAACGAAGGCGAAGGCGAAGGCGUUUGCGUGUCGAGCGA .((((..((((((((((...))))))))))..)))).....--------...(((.(((((....))..........(((((.((....)).))))).))).)))... ( -34.30) >DroSec_CAF1 59397 100 + 1 AGGGAUCUGUAUCUGUCCGCGACAGAUACUACUUCUACUAC--------CAACUCAACGGCGAAGGCGAAAACGAAGGCGAAGGCGAAGGCGUUUGCGUGUCGAGCGA .((((...(((((((((...)))))))))...)))).....--------...(((.(((((....))..........(((((.((....)).))))).))).)))... ( -32.50) >DroSim_CAF1 50473 100 + 1 AGGGAUCUGUAUCUGUCCGCGACAGAUACUACUUCUACUAC--------CAACUCAACGGCGAAGGCGAAAACGAAGGCGAAGGCGAAGGCGUUUGCGUGUCGAGCGA .((((...(((((((((...)))))))))...)))).....--------...(((.(((((....))..........(((((.((....)).))))).))).)))... ( -32.50) >DroEre_CAF1 58289 96 + 1 AGGGAUCUGUAUCUGUCCGCGACAGAUACUACUUCUACUACAGUAAAACCAAGUAAACGGCGAAG------------GCGAAGGCGAAGGCGUUUGCGUGUCGAGCGA .((.....(((((((((...)))))))))((((........))))...)).......(((((...------------(((((.((....)).))))).)))))..... ( -31.20) >consensus AGGGAUCUGUAUCUGUCCGCGACAGAUACUACUUCUACUAC________CAACUCAACGGCGAAGGCGAAAACGAAGGCGAAGGCGAAGGCGUUUGCGUGUCGAGCGA .((((...(((((((((...)))))))))...)))).....................(((((....((....))...(((((.((....)).))))).)))))..... (-27.98 = -28.10 + 0.12)


| Location | 15,991,572 – 15,991,692 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.53 |
| Mean single sequence MFE | -31.54 |
| Consensus MFE | -30.04 |
| Energy contribution | -30.08 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.13 |
| Structure conservation index | 0.95 |
| SVM decision value | 0.36 |
| SVM RNA-class probability | 0.705200 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15991572 120 + 22224390 AAAACGAAGGCGAAGGCGAAGGCGUUUGCGUGUCGAGCGACAUCAUUUGCAUAACAAUGCUGGAGAUAGAGACGAUUCAAAUUUUUGGUCUCCCAGAGCAAUUUUCACGACGAUACGAUA .........(((((.((....)).)))))((((((.((((......)))).......((((((((((.((((..........)))).))))))...))))..........)))))).... ( -34.10) >DroSec_CAF1 59457 115 + 1 AAAACGAAGGCGAAGGCGAAGGCGUUUGCGUGUCGAGCGACAUCAUUUGCAUAACAAUGCUGGAGAUAGAGACGAUUCAAAUUUUUGGUCUCCCAGAGCAAUUUUCACGACG-----AUA .........(((((.((....)).))))).(((((.((((......)))).......((((((((((.((((..........)))).))))))...)))).......)))))-----... ( -31.60) >DroSim_CAF1 50533 115 + 1 AAAACGAAGGCGAAGGCGAAGGCGUUUGCGUGUCGAGCGACAUCAUUUGCAUAACAAUGCUGGAGAUAGAGACGAUUCAAAUUUUUGGUCUCCCAGAGCAAUUUUCACGACG-----AUA .........(((((.((....)).))))).(((((.((((......)))).......((((((((((.((((..........)))).))))))...)))).......)))))-----... ( -31.60) >DroEre_CAF1 58354 106 + 1 ---------GCGAAGGCGAAGGCGUUUGCGUGUCGAGCGACAUCAUUUGCAUAACAAUGCUGGCGAUAGAGACGAUUCAAAUUUUUGGUCUCCCAGAGCAAUUUUCACGAUG-----AUA ---------(((((.((....)).)))))..(((....)))((((((.((((....))))..((....(((((.(..........).))))).....)).........))))-----)). ( -28.80) >DroYak_CAF1 42579 115 + 1 AAAACGAAGGCGAAGGCGAAGGCGUUUGCGUGUCGAGCGACAUCAUUUGCAUAACAAUGCUGGAGAUAGAGACGAUUCAAAUUUUUGGUCUCCCAGAGCAAUUUUCACGACG-----AUA .........(((((.((....)).))))).(((((.((((......)))).......((((((((((.((((..........)))).))))))...)))).......)))))-----... ( -31.60) >consensus AAAACGAAGGCGAAGGCGAAGGCGUUUGCGUGUCGAGCGACAUCAUUUGCAUAACAAUGCUGGAGAUAGAGACGAUUCAAAUUUUUGGUCUCCCAGAGCAAUUUUCACGACG_____AUA .........(((((.((....)).)))))..((((.((((......)))).......((((((((((.((((..........)))).))))))...)))).......))))......... (-30.04 = -30.08 + 0.04)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:10:59 2006