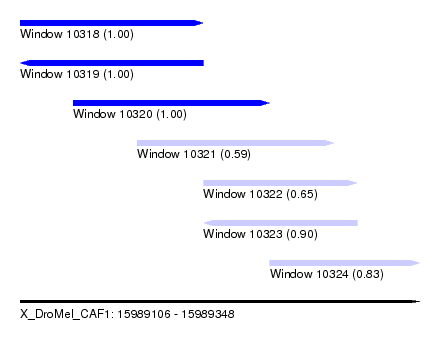
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 15,989,106 – 15,989,348 |
| Length | 242 |
| Max. P | 0.999971 |

| Location | 15,989,106 – 15,989,217 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.59 |
| Mean single sequence MFE | -33.52 |
| Consensus MFE | -30.84 |
| Energy contribution | -31.00 |
| Covariance contribution | 0.16 |
| Combinations/Pair | 1.03 |
| Mean z-score | -3.59 |
| Structure conservation index | 0.92 |
| SVM decision value | 5.06 |
| SVM RNA-class probability | 0.999971 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15989106 111 + 22224390 UCCCGCUCACUCCCGCACGGUAGA--------AUCAUCGAA-AAAGAACCAUAAAGAUGACACCACAUUGUUUUAUCUUUAUCGCAUAAUUGCGGUCAAUUCUCCUUCGAUCGCCGAGCG ...(((((.((.((....)).)).--------...((((((-..((((..((((((((((.((......)).))))))))))((((....)))).....))))..))))))....))))) ( -31.00) >DroSec_CAF1 57018 111 + 1 UCCCGCUCACUCCCGCACCGGAGA--------AUCGUCGAA-AAAGAACCAUAAAGAUGACACCACAUUGUUUUAUCUUUAUCGCAUAAUUGCGGUCAAUUCUCCUUCGAUCGCCGAGCG ...(((((.((((......)))).--------...((((((-..((((..((((((((((.((......)).))))))))))((((....)))).....))))..))))))....))))) ( -33.90) >DroSim_CAF1 48085 111 + 1 UCCCGCUCACUCCCGCACCGGAGA--------AUCGUCGAA-AGAGAACCAUAAAGAUGACACCACAUUGUUUUAUCUUUAUCGCAUAAUUGCGGUCAAUUCUCCUUCGAUCGCCGAGCG ...(((((.((((......)))).--------...((((((-.(((((..((((((((((.((......)).))))))))))((((....)))).....))))).))))))....))))) ( -37.40) >DroEre_CAF1 55876 111 + 1 UCCCGCUCACUCCCGCACCGGAGA--------GUCAUCGAA-AAAGAACCAUAAAGAUGACACCACAUUGUUUUAUCUUUAUCGCAUAAUUGCGGUCAAUUCUCCUUCGAUCGCCGAGCG ...(((((.((((......)))).--------((.((((((-..((((..((((((((((.((......)).))))))))))((((....)))).....))))..)))))).)).))))) ( -35.50) >DroYak_CAF1 39930 120 + 1 UCCCGCUCACUCCCGCACCGGAGAAUCGGAGAAUCGUCGAAAAAAGAACCAUAAAGAUGACACCACAUUGUUUUAUCUUUAUCGCAUAAUUGCGGUCAAUUCUCCUUCGAUCGCCGAACG ...((.((.((((......))))....(((((((.(.(............((((((((((.((......)).))))))))))((((....))))).).)))))))...)).))....... ( -29.80) >consensus UCCCGCUCACUCCCGCACCGGAGA________AUCGUCGAA_AAAGAACCAUAAAGAUGACACCACAUUGUUUUAUCUUUAUCGCAUAAUUGCGGUCAAUUCUCCUUCGAUCGCCGAGCG ...(((((.((((......))))............((((((...((((..((((((((((.((......)).))))))))))((((....)))).....))))..))))))....))))) (-30.84 = -31.00 + 0.16)


| Location | 15,989,106 – 15,989,217 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.59 |
| Mean single sequence MFE | -39.40 |
| Consensus MFE | -34.62 |
| Energy contribution | -34.26 |
| Covariance contribution | -0.36 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.65 |
| Structure conservation index | 0.88 |
| SVM decision value | 3.63 |
| SVM RNA-class probability | 0.999464 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15989106 111 - 22224390 CGCUCGGCGAUCGAAGGAGAAUUGACCGCAAUUAUGCGAUAAAGAUAAAACAAUGUGGUGUCAUCUUUAUGGUUCUUU-UUCGAUGAU--------UCUACCGUGCGGGAGUGAGCGGGA (((((....((((((((((((((...((((....))))((((((((...((......))...))))))))))))))))-)))))).((--------(((........))))))))))... ( -36.40) >DroSec_CAF1 57018 111 - 1 CGCUCGGCGAUCGAAGGAGAAUUGACCGCAAUUAUGCGAUAAAGAUAAAACAAUGUGGUGUCAUCUUUAUGGUUCUUU-UUCGACGAU--------UCUCCGGUGCGGGAGUGAGCGGGA ((((((.((.(((((((((((((...((((....))))((((((((...((......))...))))))))))))))))-)))))))..--------.((((......))))))))))... ( -39.60) >DroSim_CAF1 48085 111 - 1 CGCUCGGCGAUCGAAGGAGAAUUGACCGCAAUUAUGCGAUAAAGAUAAAACAAUGUGGUGUCAUCUUUAUGGUUCUCU-UUCGACGAU--------UCUCCGGUGCGGGAGUGAGCGGGA ((((((.((.(((((((((((((...((((....))))((((((((...((......))...))))))))))))))))-)))))))..--------.((((......))))))))))... ( -42.30) >DroEre_CAF1 55876 111 - 1 CGCUCGGCGAUCGAAGGAGAAUUGACCGCAAUUAUGCGAUAAAGAUAAAACAAUGUGGUGUCAUCUUUAUGGUUCUUU-UUCGAUGAC--------UCUCCGGUGCGGGAGUGAGCGGGA (((((....((((((((((((((...((((....))))((((((((...((......))...))))))))))))))))-)))))).((--------((.(((...))))))))))))... ( -41.10) >DroYak_CAF1 39930 120 - 1 CGUUCGGCGAUCGAAGGAGAAUUGACCGCAAUUAUGCGAUAAAGAUAAAACAAUGUGGUGUCAUCUUUAUGGUUCUUUUUUCGACGAUUCUCCGAUUCUCCGGUGCGGGAGUGAGCGGGA (((((((.(((((..((((((..(((((((....))).((((((((...((......))...))))))))))))..))))))..))).)).))....((((......)))).)))))... ( -37.60) >consensus CGCUCGGCGAUCGAAGGAGAAUUGACCGCAAUUAUGCGAUAAAGAUAAAACAAUGUGGUGUCAUCUUUAUGGUUCUUU_UUCGACGAU________UCUCCGGUGCGGGAGUGAGCGGGA ((((((.((.(((((((((((((...((((....))))((((((((...((......))...)))))))))))))))).)))))))...........((((......))))))))))... (-34.62 = -34.26 + -0.36)


| Location | 15,989,138 – 15,989,257 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 98.99 |
| Mean single sequence MFE | -29.18 |
| Consensus MFE | -28.86 |
| Energy contribution | -28.54 |
| Covariance contribution | -0.32 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.06 |
| Structure conservation index | 0.99 |
| SVM decision value | 2.77 |
| SVM RNA-class probability | 0.996899 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15989138 119 + 22224390 A-AAAGAACCAUAAAGAUGACACCACAUUGUUUUAUCUUUAUCGCAUAAUUGCGGUCAAUUCUCCUUCGAUCGCCGAGCGCUCUUUUGCCUCUUGACUUGGAUUUCGGCAUUGACUUUGA .-((((.(((((((((((((.((......)).)))))))))).(((....))))))..........(((((.((((((.((......)).(((......))).))))))))))))))).. ( -28.70) >DroSec_CAF1 57050 119 + 1 A-AAAGAACCAUAAAGAUGACACCACAUUGUUUUAUCUUUAUCGCAUAAUUGCGGUCAAUUCUCCUUCGAUCGCCGAGCGCUCUUUUGCCUCUUGACUUGGAUUUCGGCAUUGACUUUGA .-((((.(((((((((((((.((......)).)))))))))).(((....))))))..........(((((.((((((.((......)).(((......))).))))))))))))))).. ( -28.70) >DroSim_CAF1 48117 119 + 1 A-AGAGAACCAUAAAGAUGACACCACAUUGUUUUAUCUUUAUCGCAUAAUUGCGGUCAAUUCUCCUUCGAUCGCCGAGCGCUCUUUUGCCUCUUGACUUGGAUUUCGGCAUUGACUUUGA .-.(((((..((((((((((.((......)).))))))))))((((....)))).....)))))..(((((.((((((.((......)).(((......))).)))))))))))...... ( -30.80) >DroEre_CAF1 55908 119 + 1 A-AAAGAACCAUAAAGAUGACACCACAUUGUUUUAUCUUUAUCGCAUAAUUGCGGUCAAUUCUCCUUCGAUCGCCGAGCGCUCUUUUGCCUCUUGACUUGGAUUUCGGCAUUGACUUUGA .-((((.(((((((((((((.((......)).)))))))))).(((....))))))..........(((((.((((((.((......)).(((......))).))))))))))))))).. ( -28.70) >DroYak_CAF1 39970 120 + 1 AAAAAGAACCAUAAAGAUGACACCACAUUGUUUUAUCUUUAUCGCAUAAUUGCGGUCAAUUCUCCUUCGAUCGCCGAACGCUCUUUUGCCUCUUGACUUGGAUUUCGGCAUUGACUUUGA ..((((.(((((((((((((.((......)).)))))))))).(((....))))))..........(((((.((((((.((......)).(((......))).))))))))))))))).. ( -29.00) >consensus A_AAAGAACCAUAAAGAUGACACCACAUUGUUUUAUCUUUAUCGCAUAAUUGCGGUCAAUUCUCCUUCGAUCGCCGAGCGCUCUUUUGCCUCUUGACUUGGAUUUCGGCAUUGACUUUGA ..((((.(((((((((((((.((......)).)))))))))).(((....))))))..........(((((.((((((.((......)).(((......))).))))))))))))))).. (-28.86 = -28.54 + -0.32)

| Location | 15,989,177 – 15,989,296 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 98.99 |
| Mean single sequence MFE | -33.56 |
| Consensus MFE | -30.34 |
| Energy contribution | -30.74 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.39 |
| Structure conservation index | 0.90 |
| SVM decision value | 0.11 |
| SVM RNA-class probability | 0.588514 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15989177 119 + 22224390 AUCGCAUAAUUGCGGUCAAUUCUCCUUCGAUCGCCGAGCGCUCUUUUGCCUCUUGACUUGGAUUUCGGCAUUGACUUUGAUCAUUUGCCGUGGCCACAUUCACCAGGGCUCG-AACUCGA ((((((....)))))).........(((((..((((((.((......))))).(((..(((....(((((.(((......)))..)))))...)))...)))....))))))-))..... ( -33.30) >DroSec_CAF1 57089 119 + 1 AUCGCAUAAUUGCGGUCAAUUCUCCUUCGAUCGCCGAGCGCUCUUUUGCCUCUUGACUUGGAUUUCGGCAUUGACUUUGAUCAUUUGCCGUGGCCACAUUCACCAGGGCUCG-AACUCGA ((((((....)))))).........(((((..((((((.((......))))).(((..(((....(((((.(((......)))..)))))...)))...)))....))))))-))..... ( -33.30) >DroSim_CAF1 48156 119 + 1 AUCGCAUAAUUGCGGUCAAUUCUCCUUCGAUCGCCGAGCGCUCUUUUGCCUCUUGACUUGGAUUUCGGCAUUGACUUUGAUCAUUUGCCGUGGCCACAUUCACCAGGGCUCG-AACUCGA ((((((....)))))).........(((((..((((((.((......))))).(((..(((....(((((.(((......)))..)))))...)))...)))....))))))-))..... ( -33.30) >DroEre_CAF1 55947 120 + 1 AUCGCAUAAUUGCGGUCAAUUCUCCUUCGAUCGCCGAGCGCUCUUUUGCCUCUUGACUUGGAUUUCGGCAUUGACUUUGAUCAUUUGCCGUGGCCACAUUCACCAGGGCUCGGAACUCGA ((((((....))))))..........((((...((((((((......)).......(((((....(((((.(((......)))..)))))((....))....)))))))))))...)))) ( -37.50) >DroYak_CAF1 40010 119 + 1 AUCGCAUAAUUGCGGUCAAUUCUCCUUCGAUCGCCGAACGCUCUUUUGCCUCUUGACUUGGAUUUCGGCAUUGACUUUGAUCAUUUGCCGUGGCCACAUUCACCAGGGCUCG-AACUCUA ...(((....)))((((((.......(((((.((((((.((......)).(((......))).)))))))))))..))))))....(((.(((.........))).)))...-....... ( -30.40) >consensus AUCGCAUAAUUGCGGUCAAUUCUCCUUCGAUCGCCGAGCGCUCUUUUGCCUCUUGACUUGGAUUUCGGCAUUGACUUUGAUCAUUUGCCGUGGCCACAUUCACCAGGGCUCG_AACUCGA ((((((....))))))..........((((....(((((((......)).......(((((....(((((.(((......)))..)))))((....))....))))))))))....)))) (-30.34 = -30.74 + 0.40)



| Location | 15,989,217 – 15,989,310 |
|---|---|
| Length | 93 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 82.17 |
| Mean single sequence MFE | -27.97 |
| Consensus MFE | -19.62 |
| Energy contribution | -20.22 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.70 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.23 |
| SVM RNA-class probability | 0.645861 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15989217 93 + 22224390 CUCUUUUGCCUCUUGACUUGGAUUUCGGCAUUGACUUUGAUCAUUUGCCGUGGCCACAUUCACCAGGGCUCG-AACUCGAGCU--------------------------CGAUCAACUUG .((...(((((((......)))....))))..))..((((((.......((((......))))..(((((((-....))))))--------------------------))))))).... ( -26.00) >DroSec_CAF1 57129 93 + 1 CUCUUUUGCCUCUUGACUUGGAUUUCGGCAUUGACUUUGAUCAUUUGCCGUGGCCACAUUCACCAGGGCUCG-AACUCGAGCC--------------------------CGAUCGACUUG .((...(((((((......)))....))))..))..((((((.......((((......))))..(((((((-....))))))--------------------------))))))).... ( -28.70) >DroSim_CAF1 48196 93 + 1 CUCUUUUGCCUCUUGACUUGGAUUUCGGCAUUGACUUUGAUCAUUUGCCGUGGCCACAUUCACCAGGGCUCG-AACUCGAGCC--------------------------CGAUCGACUUG .((...(((((((......)))....))))..))..((((((.......((((......))))..(((((((-....))))))--------------------------))))))).... ( -28.70) >DroEre_CAF1 55987 120 + 1 CUCUUUUGCCUCUUGACUUGGAUUUCGGCAUUGACUUUGAUCAUUUGCCGUGGCCACAUUCACCAGGGCUCGGAACUCGAACUCCAACUCCAACUCACACUCGCACUCGAUAUCGGCUUG .......(((...(((.(((((...(((((.(((......)))..))))).((((.(........))))).(((........)))...))))).)))...(((....)))....)))... ( -26.60) >DroYak_CAF1 40050 119 + 1 CUCUUUUGCCUCUUGACUUGGAUUUCGGCAUUGACUUUGAUCAUUUGCCGUGGCCACAUUCACCAGGGCUCG-AACUCUAACUCCAACUCCAACUCCAACUCGAGCUCUCGAUCGGCUUG .......(((...(((..(((....(((((.(((......)))..)))))...)))...)))(.((((((((-(..........................))))))))).)...)))... ( -29.87) >consensus CUCUUUUGCCUCUUGACUUGGAUUUCGGCAUUGACUUUGAUCAUUUGCCGUGGCCACAUUCACCAGGGCUCG_AACUCGAGCU__________________________CGAUCGACUUG ........(((..(((..(((....(((((.(((......)))..)))))...)))...)))..)))(((((.....)))))...................................... (-19.62 = -20.22 + 0.60)


| Location | 15,989,217 – 15,989,310 |
|---|---|
| Length | 93 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 82.17 |
| Mean single sequence MFE | -32.64 |
| Consensus MFE | -23.30 |
| Energy contribution | -23.26 |
| Covariance contribution | -0.04 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.11 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.99 |
| SVM RNA-class probability | 0.896251 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15989217 93 - 22224390 CAAGUUGAUCG--------------------------AGCUCGAGUU-CGAGCCCUGGUGAAUGUGGCCACGGCAAAUGAUCAAAGUCAAUGCCGAAAUCCAAGUCAAGAGGCAAAAGAG ........(((--------------------------(((....)))-)))(((((..(((...(((...(((((..((((....)))).)))))....)))..))))).)))....... ( -26.90) >DroSec_CAF1 57129 93 - 1 CAAGUCGAUCG--------------------------GGCUCGAGUU-CGAGCCCUGGUGAAUGUGGCCACGGCAAAUGAUCAAAGUCAAUGCCGAAAUCCAAGUCAAGAGGCAAAAGAG ...(((....(--------------------------((((((....-)))))))...(((...(((...(((((..((((....)))).)))))....)))..)))...)))....... ( -32.10) >DroSim_CAF1 48196 93 - 1 CAAGUCGAUCG--------------------------GGCUCGAGUU-CGAGCCCUGGUGAAUGUGGCCACGGCAAAUGAUCAAAGUCAAUGCCGAAAUCCAAGUCAAGAGGCAAAAGAG ...(((....(--------------------------((((((....-)))))))...(((...(((...(((((..((((....)))).)))))....)))..)))...)))....... ( -32.10) >DroEre_CAF1 55987 120 - 1 CAAGCCGAUAUCGAGUGCGAGUGUGAGUUGGAGUUGGAGUUCGAGUUCCGAGCCCUGGUGAAUGUGGCCACGGCAAAUGAUCAAAGUCAAUGCCGAAAUCCAAGUCAAGAGGCAAAAGAG ...(((....(((....)))...(((.(((((...((.(((((.....)))))))((((.......))))(((((..((((....)))).)))))...))))).)))...)))....... ( -36.30) >DroYak_CAF1 40050 119 - 1 CAAGCCGAUCGAGAGCUCGAGUUGGAGUUGGAGUUGGAGUUAGAGUU-CGAGCCCUGGUGAAUGUGGCCACGGCAAAUGAUCAAAGUCAAUGCCGAAAUCCAAGUCAAGAGGCAAAAGAG ...(((((.(....).))((.(((((.((((.(((((..((.((...-...(((.((((.......)))).)))......))))..))))).))))..))))).))....)))....... ( -35.80) >consensus CAAGUCGAUCG__________________________AGCUCGAGUU_CGAGCCCUGGUGAAUGUGGCCACGGCAAAUGAUCAAAGUCAAUGCCGAAAUCCAAGUCAAGAGGCAAAAGAG ......................................(((((.....)))))(((..(((...(((...(((((..((((....)))).)))))....)))..)))..)))........ (-23.30 = -23.26 + -0.04)



| Location | 15,989,257 – 15,989,348 |
|---|---|
| Length | 91 |
| Sequences | 4 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 80.60 |
| Mean single sequence MFE | -37.12 |
| Consensus MFE | -27.20 |
| Energy contribution | -28.20 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.85 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.73 |
| SVM RNA-class probability | 0.834242 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15989257 91 + 22224390 UCAUUUGCCGUGGCCACAUUCACCAGGGCUCGAACUCGAGCU--------------------------CGAUCAACUUGGAUCGAGACGAACGGGGGAGGGCUAUAGCCAUGGCU--AU ......((((((((..(........)(((((...((((.(((--------------------------(((((......))))))).)...))))...)))))...)))))))).--.. ( -36.30) >DroSec_CAF1 57169 93 + 1 UCAUUUGCCGUGGCCACAUUCACCAGGGCUCGAACUCGAGCC--------------------------CGAUCGACUUGGCUCGAGACGGACGGGGGAGGGCUAUAGCAAUGGCUAUAU (((((.((..(((((...(((.((.(((((((....))))))--------------------------)(.(((.((((...)))).))).))).))).)))))..)))))))...... ( -37.90) >DroSim_CAF1 48236 93 + 1 UCAUUUGCCGUGGCCACAUUCACCAGGGCUCGAACUCGAGCC--------------------------CGAUCGACUUGGCUCGAGACGGACGGGGGAGGGCUAUAGCCAUGGCUCUAU ......((((((((....(((.((.(((((((....))))))--------------------------)(.(((.((((...)))).))).)))..))).......))))))))..... ( -37.50) >DroYak_CAF1 40090 115 + 1 UCAUUUGCCGUGGCCACAUUCACCAGGGCUCGAACUCUAACUCCAACUCCAACUCCAACUCGAGCUCUCGAUCGGCUUGGAUCG--ACGAACGGGGGAGGGCUAUUGCCAUGGCU--AU ......((((((((....(((.((.((((((((..........................))))))))((((((......)))))--).....))..))).......)))))))).--.. ( -36.77) >consensus UCAUUUGCCGUGGCCACAUUCACCAGGGCUCGAACUCGAGCC__________________________CGAUCGACUUGGAUCGAGACGAACGGGGGAGGGCUAUAGCCAUGGCU__AU ......((((((((....(((.((..((((((....))))))..........................(((((......)))))........))..))).......))))))))..... (-27.20 = -28.20 + 1.00)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:10:52 2006