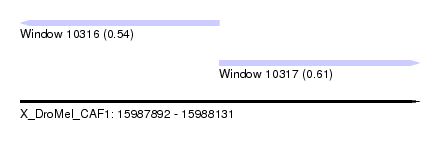
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 15,987,892 – 15,988,131 |
| Length | 239 |
| Max. P | 0.609932 |

| Location | 15,987,892 – 15,988,011 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.91 |
| Mean single sequence MFE | -25.16 |
| Consensus MFE | -23.58 |
| Energy contribution | -24.22 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.13 |
| Structure conservation index | 0.94 |
| SVM decision value | 0.02 |
| SVM RNA-class probability | 0.542910 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15987892 119 - 22224390 CGCCCAUCGCCUUGACUAUAUCGGUUCCACAUUAAGCACAAUUAGUGUGGCCGUGCCGUCAAUUAAUUUAUCUUCUGUUAAUUACACUCGUUAUUUGUAA-AACAUAAGGGUAUAUUGCA .((.....((((((((.....(((..(((((((((......))))))))))))....))))).........(((.((((..(((((.........)))))-)))).)))))).....)). ( -27.10) >DroSec_CAF1 55821 120 - 1 CGCCCAUCGCCUUGACUAUAUCGGUUCCACAUUAAGCACAAUUAGUGUGGCCGUGCCGUCAAUUAAUUUAUCUUCUGUUAAUUAUACUCGUUAUUUGUAAAAACAUAAGGGUAUAUUGCA ........((.(((((.....(((..(((((((((......))))))))))))....)))))....................(((((((......(((....)))...)))))))..)). ( -25.90) >DroSim_CAF1 46913 120 - 1 CGCCCAUCGCCUUGACUAUAUCGGUUCCACAUUAAGCACAAUUAGUGUGGCCGUGCCGUCAAUUAAUUUAUCUUCUGUUAAUUACACUCGUUAUUUGUAAAAACAUAAGGGUAUAUUGCA .((.....((((((((.....(((..(((((((((......))))))))))))....))))).........(((.((((..(((((.........))))).)))).)))))).....)). ( -27.00) >DroEre_CAF1 54651 119 - 1 CGCCCAUCGCCUGGACUAUAUCGGUUCCACAUUAAGCACAAUUAGUUUGGCCGUGCCGUCAAUUAAUUUAUCUUCUGUUAAUUACACUCGUUAUUUGUAA-AACAUAAGGGUAUAUGGCA .(((....(((..(((.....(((..(((.(((((......))))).))))))....)))...........(((.((((..(((((.........)))))-)))).))))))....))). ( -23.10) >DroYak_CAF1 38628 119 - 1 CGCCCAUCGCCUUGACUAUAUCGGUUCCACAUUAAGCACAAUUUGUGUGGCCGUGCCGUCAAUUAAUUUAUCUACUGUUAAUUACACUCGUUAUUUGUUA-AACAUAAGGGUAUAUCGCA .((((......(((((.....((((..((((............))))..))))....)))))(((((..((..(((((.....)))...)).))..))))-)......))))........ ( -22.70) >consensus CGCCCAUCGCCUUGACUAUAUCGGUUCCACAUUAAGCACAAUUAGUGUGGCCGUGCCGUCAAUUAAUUUAUCUUCUGUUAAUUACACUCGUUAUUUGUAA_AACAUAAGGGUAUAUUGCA .((((......(((((.....(((..(((((((((......))))))))))))....))))).............((((...((((.........))))..))))...))))........ (-23.58 = -24.22 + 0.64)


| Location | 15,988,011 – 15,988,131 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.83 |
| Mean single sequence MFE | -28.80 |
| Consensus MFE | -24.96 |
| Energy contribution | -25.00 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.25 |
| Structure conservation index | 0.87 |
| SVM decision value | 0.15 |
| SVM RNA-class probability | 0.609932 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15988011 120 + 22224390 CAAUUUUGAUAAGCGAUUAAACGAUUACCCAUUAGUCGCGACUUAACGGUCGCAGGCCCCUUUUCGUAUCUCUUAGUCCUUGGAUCUUGGAUCUACCAAGGGAUCUGUACACGAAACCCA ...(((((....((((((((...........))))))))((((....))))((((..(((((...(((..(((.((((....)).)).)))..))).)))))..))))...))))).... ( -31.90) >DroSec_CAF1 55941 120 + 1 CAAUUUUGAUAAGCGAUUAAACGAUUACCCAUUAGUCGCGACUUAACGGUCGCAGGCCCCUUUUCGUAUCUCUUUGUCCUUGGAUCUUGGAUCUACCAAGGGAUCACUACACGAAACGCA ............(((.......(((((.....)))))((((((....)))))).(..(((((...(((..(((..(((....)))...)))..))).)))))..)...........))). ( -29.20) >DroSim_CAF1 47033 120 + 1 CAAUUUUGAUAAGCGAUUAAACGAUUACCCAUUAGUCGCGACUUAACGGUCGCAGGCCCCUUUUCGUAUCUCUUUGUCCUUGGAUCUUGGAUCUACCAAGGGAUCACUACACGAAACGCA ............(((.......(((((.....)))))((((((....)))))).(..(((((...(((..(((..(((....)))...)))..))).)))))..)...........))). ( -29.20) >DroEre_CAF1 54770 118 + 1 CAAUUUUGAUAAGCGAUUAAACGAUUAC-CAUUAGUCGCGACUUAACGGUCCCAGGCCCCUU-UUGUAUCUCUUUGUCCUCGGAUCUUGAAUCUACCAAGGGAUCCCGAGCCGAAACCCA ............((((((((........-..)))))))).......(((..(..(..(((((-..(((..((...(((....)))...))...))).)))))..)..)..)))....... ( -25.50) >DroYak_CAF1 38747 120 + 1 CAAUUUUGAUAAGCGAUUAAACGAUUACCCAUUAGUCGCGACUUAACGGUCCCAGGCCCCUUUUUGUAUCUCCUUGUCCACGGAUCUUGGAUCUACCAAGGGAUCUCUACCCGAAACCCA ...(((((....((((((((...........))))))))........(((...((..(((((...(((..(((..(((....)))...)))..))).)))))..))..)))))))).... ( -28.20) >consensus CAAUUUUGAUAAGCGAUUAAACGAUUACCCAUUAGUCGCGACUUAACGGUCGCAGGCCCCUUUUCGUAUCUCUUUGUCCUUGGAUCUUGGAUCUACCAAGGGAUCACUACACGAAACCCA ...(((((....((((((((...........))))))))((((....))))...(..(((((...(((..(((..(((....)))...)))..))).)))))..)......))))).... (-24.96 = -25.00 + 0.04)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:10:46 2006