
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 15,987,637 – 15,987,742 |
| Length | 105 |
| Max. P | 0.857061 |

| Location | 15,987,637 – 15,987,742 |
|---|---|
| Length | 105 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 76.01 |
| Mean single sequence MFE | -22.96 |
| Consensus MFE | -4.25 |
| Energy contribution | -4.37 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.30 |
| Mean z-score | -1.87 |
| Structure conservation index | 0.19 |
| SVM decision value | 0.81 |
| SVM RNA-class probability | 0.857061 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15987637 105 + 22224390 AAAGUAAA------------UGCACUUUUUUCACAGUGAAUUUGAAACUUAAUUGCAGAU-GAUAGCAUUAAAAAGUG--CCAAAGUAUAGGCAGUGCAAUAUACUGUGAAUUAGAUGAC ........------------..((((...(((((((((...(((.......(((((..((-(...(((((....))))--).....)))..))))).)))..)))))))))..)).)).. ( -21.90) >DroSec_CAF1 55563 105 + 1 AAAGUAAA------------UGCACUUUUUGUACAGUGCAUUUGAAACUUAAUUGCAGAU-GAUAGCAUUAAAAAGUG--CCAAAGUAUAGGCUUUGCAAUGAACUGUGAAUUAGAUGAC ....((((------------((((((........))))))))))....((((((((((..-....(((((....))))--)((((((....)))))).......)))).))))))..... ( -24.50) >DroSim_CAF1 46662 94 + 1 AGAGUAAA------------UGCACUUUUUGUACAGUGCAUUUGAAACUUAAUUGCAGAU-GAUAGCAUUAAAAAGUG--CCAAAGUAUAGGUCUUGCAAU-----------AAGAUGAC ....((((------------((((((........))))))))))...((((.((((((((-.((((((((....))))--).....)))..))).))))))-----------)))..... ( -22.60) >DroEre_CAF1 54406 120 + 1 AGAGUAAAAUCUAUAUGCUAUGUUUUUUUUUCACAGUGAGUUUGAAACUUAAUUGCAGAUAGAUAACAUUAAAAAGUCCCCCAAAGUAUAGGUUUUGGAAUGGACAGUGAACUAAAUGAC ........((((((.(((...(((((...(((.....)))...)))))......))).))))))..((((.....((((.((((((......))))))...))))((....)).)))).. ( -24.20) >DroYak_CAF1 38361 111 + 1 AGAGUAAAAUCUGUUUGCUAUGCACUUUUUUCACAGUGCAAUUGAAACUUAAUUGCAGAUAGAUAACAUUCAAAAGUC--CCAAAGUAUAGGUUUAGGUUU-------AAUUGAAAUUAC ...((((.((((((((((...(((((........)))))((((((...))))))))))))))))....(((((..(..--((.(((......))).))..)-------..))))).)))) ( -21.60) >consensus AGAGUAAA____________UGCACUUUUUUCACAGUGCAUUUGAAACUUAAUUGCAGAU_GAUAGCAUUAAAAAGUG__CCAAAGUAUAGGUUUUGCAAU__AC_GUGAAUUAGAUGAC ...((((.............((((((........))))))............))))................................................................ ( -4.25 = -4.37 + 0.12)


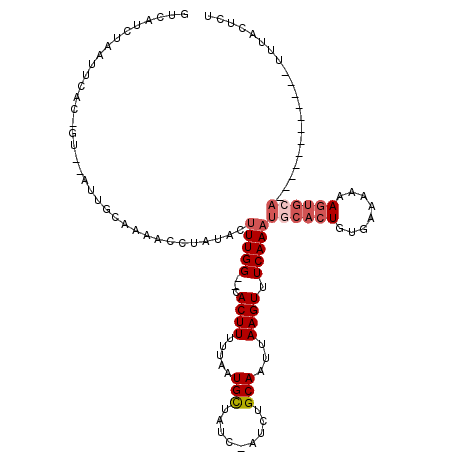
| Location | 15,987,637 – 15,987,742 |
|---|---|
| Length | 105 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 76.01 |
| Mean single sequence MFE | -19.55 |
| Consensus MFE | -6.08 |
| Energy contribution | -7.24 |
| Covariance contribution | 1.16 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.96 |
| Structure conservation index | 0.31 |
| SVM decision value | 0.69 |
| SVM RNA-class probability | 0.823424 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15987637 105 - 22224390 GUCAUCUAAUUCACAGUAUAUUGCACUGCCUAUACUUUGG--CACUUUUUAAUGCUAUC-AUCUGCAAUUAAGUUUCAAAUUCACUGUGAAAAAAGUGCA------------UUUACUUU .........((((((((..(((....((((........))--))...((((((((....-....)).)))))).....)))..)))))))).((((((..------------..)))))) ( -20.60) >DroSec_CAF1 55563 105 - 1 GUCAUCUAAUUCACAGUUCAUUGCAAAGCCUAUACUUUGG--CACUUUUUAAUGCUAUC-AUCUGCAAUUAAGUUUCAAAUGCACUGUACAAAAAGUGCA------------UUUACUUU ...................((((((..(((........))--)........(((....)-)).)))))).((((...(((((((((........))))))------------))))))). ( -19.70) >DroSim_CAF1 46662 94 - 1 GUCAUCUU-----------AUUGCAAGACCUAUACUUUGG--CACUUUUUAAUGCUAUC-AUCUGCAAUUAAGUUUCAAAUGCACUGUACAAAAAGUGCA------------UUUACUCU .....(((-----------((((((.((.........(((--((........)))))..-.))))))).))))....(((((((((........))))))------------)))..... ( -19.30) >DroEre_CAF1 54406 120 - 1 GUCAUUUAGUUCACUGUCCAUUCCAAAACCUAUACUUUGGGGGACUUUUUAAUGUUAUCUAUCUGCAAUUAAGUUUCAAACUCACUGUGAAAAAAAAACAUAGCAUAUAGAUUUUACUCU ...............((((...(((((........))))).))))........((.((((((.(((.......(((((.........)))))..........))).))))))...))... ( -18.13) >DroYak_CAF1 38361 111 - 1 GUAAUUUCAAUU-------AAACCUAAACCUAUACUUUGG--GACUUUUGAAUGUUAUCUAUCUGCAAUUAAGUUUCAAUUGCACUGUGAAAAAAGUGCAUAGCAAACAGAUUUUACUCU ((((..((....-------...((((((.......)))))--)....((((.(((.........))).))))((((....((((((........))))))....)))).))..))))... ( -20.00) >consensus GUCAUCUAAUUCAC_GU__AUUGCAAAACCUAUACUUUGG__CACUUUUUAAUGCUAUC_AUCUGCAAUUAAGUUUCAAAUGCACUGUGAAAAAAGUGCA____________UUUACUCU ...................................(((((...((((.....(((.........)))...)))).)))))((((((........)))))).................... ( -6.08 = -7.24 + 1.16)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:10:44 2006