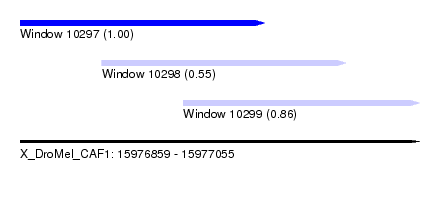
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 15,976,859 – 15,977,055 |
| Length | 196 |
| Max. P | 0.999565 |

| Location | 15,976,859 – 15,976,979 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 98.33 |
| Mean single sequence MFE | -46.84 |
| Consensus MFE | -46.72 |
| Energy contribution | -46.56 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.33 |
| Structure conservation index | 1.00 |
| SVM decision value | 3.73 |
| SVM RNA-class probability | 0.999565 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
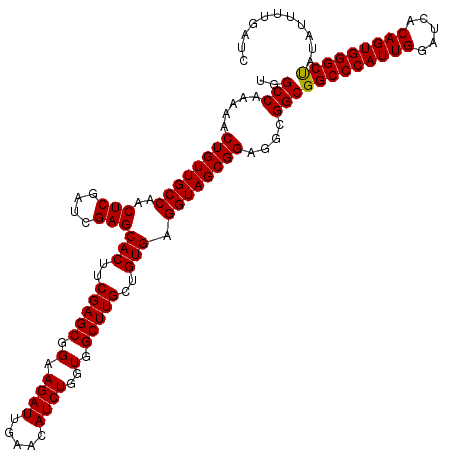
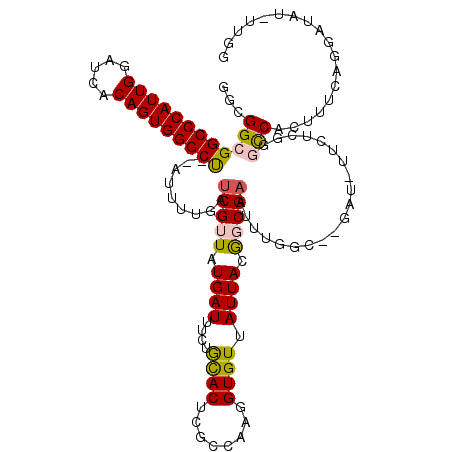
>X_DroMel_CAF1 15976859 120 + 22224390 UGGCCAAAAACUGUUGCCAACUCGAUCGAGCACUUCGAGCGGAAGAUUGAACAUCUGGUGGCUUGCUGUGAGGUAGCGGAGGCGGCGGCCCAUUGGAUCACAGUGGGCCAUAUUUUGAUC ..(((.....((((((((..(((....)))(((..(((((.(.((((.....))))..).)))))..))).))))))))....)))(((((((((.....)))))))))........... ( -49.00) >DroSec_CAF1 44659 120 + 1 UGGCCAAAAACUGUUGCCAACUCGAUCGAGCACUUCGAGCGGAAGAUUGAACAUCUGGUGGCUUGCUGUGAGGUAGCGGAGGCGGCGGCCCAUUGGAUCACAGUGGGCUAUAUUUUGAUC ..(((.....((((((((..(((....)))(((..(((((.(.((((.....))))..).)))))..))).))))))))....)))(((((((((.....)))))))))........... ( -46.30) >DroSim_CAF1 35784 120 + 1 UGGCCAAAAACUGUUGCCAACUCGAUCGAGCACUUCGAGCGGAAGAUUGAACAUCUGGUGGCUUGCUGUGAGGUAGCGGAGGCGGCGGCCCAUUGGAUCACAGUGGGCUAUAUUUUGAUC ..(((.....((((((((..(((....)))(((..(((((.(.((((.....))))..).)))))..))).))))))))....)))(((((((((.....)))))))))........... ( -46.30) >DroEre_CAF1 43552 118 + 1 UGGCCAAAAACUGUUGCCAACUCGAUCGAGCACUUCGAGCGGAAGAUUGAACAUCUGGUGGCUUGCUGUGAGGUAGCGGAGGCGGCGGCCCAUUGGAUCACAGUGGGCU--AUUUCGAUC ..(((.....((((((((..(((....)))(((..(((((.(.((((.....))))..).)))))..))).))))))))....)))(((((((((.....)))))))))--......... ( -46.30) >DroYak_CAF1 27411 118 + 1 UGGCCAAAAACUGUUGCCAACUCGAUCGAGCACUUCGAGCGGAAGAUUGAACAUCUGGUGGCUUGCUGUGAGGUAGCGGAGGCGGCGGCCCAUUGGAUCACAGUGGGCU--AUUUUGAUC ..(((.....((((((((..(((....)))(((..(((((.(.((((.....))))..).)))))..))).))))))))....)))(((((((((.....)))))))))--......... ( -46.30) >consensus UGGCCAAAAACUGUUGCCAACUCGAUCGAGCACUUCGAGCGGAAGAUUGAACAUCUGGUGGCUUGCUGUGAGGUAGCGGAGGCGGCGGCCCAUUGGAUCACAGUGGGCUAUAUUUUGAUC ..(((.....((((((((..(((....)))(((..(((((.(.((((.....))))..).)))))..))).))))))))....)))(((((((((.....)))))))))........... (-46.72 = -46.56 + -0.16)


| Location | 15,976,899 – 15,977,019 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 82.47 |
| Mean single sequence MFE | -38.20 |
| Consensus MFE | -32.48 |
| Energy contribution | -32.12 |
| Covariance contribution | -0.36 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.45 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.03 |
| SVM RNA-class probability | 0.547453 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15976899 120 + 22224390 GGAAGAUUGAACAUCUGGUGGCUUGCUGUGAGGUAGCGGAGGCGGCGGCCCAUUGGAUCACAGUGGGCCAUAUUUUGAUCGUUAUGAUUUCUAUACUCGCCAAGGUGUUAUUACAUCGAA ....(((..(((((((....((((.((((......))))))))((((((((((((.....))))))))).(((...((((.....))))...)))...))).))))))).....)))... ( -40.70) >DroSec_CAF1 44699 120 + 1 GGAAGAUUGAACAUCUGGUGGCUUGCUGUGAGGUAGCGGAGGCGGCGGCCCAUUGGAUCACAGUGGGCUAUAUUUUGAUCGUUAUGAUUUCUGCACUCGCCAAUGUGUUAUUACGUCGUA .((......(((((....((((.(((((.....)))))(((((((.(((((((((.....))))))))).......((((.....)))).)))).)))))))..)))))......))... ( -38.60) >DroSim_CAF1 35824 120 + 1 GGAAGAUUGAACAUCUGGUGGCUUGCUGUGAGGUAGCGGAGGCGGCGGCCCAUUGGAUCACAGUGGGCUAUAUUUUGAUCNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNN ....(((..((......((.((((.((((......)))))))).))(((((((((.....)))))))))....))..)))........................................ ( -30.00) >DroEre_CAF1 43592 118 + 1 GGAAGAUUGAACAUCUGGUGGCUUGCUGUGAGGUAGCGGAGGCGGCGGCCCAUUGGAUCACAGUGGGCU--AUUUCGAUCGUUAUGAUUUCUGCACUCCCGAAGGUGUUAUUACGGCGAA ....(((((((......((.((((.((((......)))))))).))(((((((((.....)))))))))--..)))))))((..((((....(((((......))))).))))..))... ( -42.50) >DroYak_CAF1 27451 118 + 1 GGAAGAUUGAACAUCUGGUGGCUUGCUGUGAGGUAGCGGAGGCGGCGGCCCAUUGGAUCACAGUGGGCU--AUUUUGAUCGUUAUGAUUUCUGCACACUCCAAGGUGUUAUUACGGCGAA ....(((..((......((.((((.((((......)))))))).))(((((((((.....)))))))))--..))..)))((..((((....((((........)))).))))..))... ( -39.20) >consensus GGAAGAUUGAACAUCUGGUGGCUUGCUGUGAGGUAGCGGAGGCGGCGGCCCAUUGGAUCACAGUGGGCUAUAUUUUGAUCGUUAUGAUUUCUGCACUCGCCAAGGUGUUAUUACGGCGAA ....(((((((......((.((((.((((......)))))))).))(((((((((.....)))))))))....)))))))............((((........))))............ (-32.48 = -32.12 + -0.36)

| Location | 15,976,939 – 15,977,055 |
|---|---|
| Length | 116 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 81.16 |
| Mean single sequence MFE | -37.11 |
| Consensus MFE | -24.99 |
| Energy contribution | -25.30 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.20 |
| Mean z-score | -2.01 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.82 |
| SVM RNA-class probability | 0.857837 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
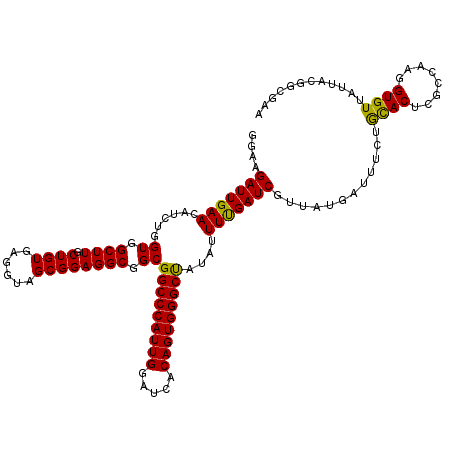
>X_DroMel_CAF1 15976939 116 + 22224390 GGCGGCGGCCCAUUGGAUCACAGUGGGCCAUAUUUUGAUCGUUAUGAUUUCUAUACUCGCCAAGGUGUUAUUACAUCGAAUUUGGC--GAU-UUCUCGUGCCACAUUCAGGAUAU-UUUG (((((.(((((((((.....))))))))).(((...((((.....))))...))).)))))..(((((....)))))((((.((((--(..-......))))).)))).......-.... ( -41.00) >DroSec_CAF1 44739 117 + 1 GGCGGCGGCCCAUUGGAUCACAGUGGGCUAUAUUUUGAUCGUUAUGAUUUCUGCACUCGCCAAUGUGUUAUUACGUCGUAAUGUGU--UUUAUUUUUUCUCCACUUUCAGGAUAU-UUGU ......(((((((((.....)))))))))(((((((((..((((((((....((((........))))......))))))))(((.--.............)))..)))))))))-.... ( -30.84) >DroEre_CAF1 43632 114 + 1 GGCGGCGGCCCAUUGGAUCACAGUGGGCU--AUUUCGAUCGUUAUGAUUUCUGCACUCCCGAAGGUGUUAUUACGGCGAACUUGCC--GAU-UACUCGGUUCACUUUCUGGUUGC-UUGG (((((((((((((((.....)))))))))--.....((((((..((((....(((((......))))).))))..))))....(((--((.-...)))))))........)))))-)... ( -39.40) >DroYak_CAF1 27491 117 + 1 GGCGGCGGCCCAUUGGAUCACAGUGGGCU--AUUUUGAUCGUUAUGAUUUCUGCACACUCCAAGGUGUUAUUACGGCGAAUUUGGCUGGUU-UUCCUGGGCCACUUUCAGCAUGUUUUGG (((((((((((((((.....)))))))))--....(((((((..((((....((((........)))).))))..))))...(((((((..-..))..)))))...))))).)))).... ( -37.20) >consensus GGCGGCGGCCCAUUGGAUCACAGUGGGCU__AUUUUGAUCGUUAUGAUUUCUGCACUCGCCAAGGUGUUAUUACGGCGAAUUUGGC__GAU_UUCUCGGGCCACUUUCAGGAUAU_UUGG ...((((((((((((.....))))))))).........(((((.((((....((((........)))).)))).)))))....................))).................. (-24.99 = -25.30 + 0.31)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:10:29 2006