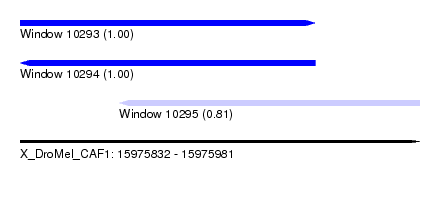
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 15,975,832 – 15,975,981 |
| Length | 149 |
| Max. P | 0.999987 |

| Location | 15,975,832 – 15,975,942 |
|---|---|
| Length | 110 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.16 |
| Mean single sequence MFE | -32.42 |
| Consensus MFE | -32.24 |
| Energy contribution | -31.76 |
| Covariance contribution | -0.48 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.46 |
| Structure conservation index | 0.99 |
| SVM decision value | 5.06 |
| SVM RNA-class probability | 0.999971 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15975832 110 + 22224390 CC---AGCCAAUCUCGAGGCCGUUAAAGAUCUUUUGAGAUCUG-------AGAUCUCUGACCAGCCAGAUGGCAAGGUCGAAACCAUGCCUUAAUUAUUAAUACACAUAAGGCUAUGUAA ..---((((.((((...(((.((((.(((((((..(....).)-------)))))).))))..)))))))((((.(((....))).))))....................))))...... ( -31.20) >DroSec_CAF1 43643 110 + 1 CU---AGCCAAUCUCGAGGCCGUGAAAGAUCUUUUGAGAUCUG-------AGAUCUCUGACCAGCCAGACGGCAAGGUCGACACCAUGCCUUAAUUAUUAAUACACAUAAGGCUAUGUAA .(---((((......(((((.(((...((((((..((((((..-------.))))))......(((....)))))))))..)))...)))))..((((........)))))))))..... ( -31.00) >DroSim_CAF1 34779 110 + 1 AU---AGCCAAUCUCGAGGCCGUGAAAGAUCUUUUGAGAUCUG-------AGAUCUCUGACCAGCCAGACGGCAAGGUCGACACCAUGCCUUAAUUAUUAAUACACAUAAGGCUAUGUAA ((---((((......(((((.(((...((((((..((((((..-------.))))))......(((....)))))))))..)))...)))))..((((........)))))))))).... ( -32.10) >DroEre_CAF1 42513 113 + 1 CUCGUGGCCAAUCUCGAGGCCGUGAAAGAUCUUUUGAGAUCUG-------AGAUCUCUGACCAGCCACCCGGCAAGGUCGACACUAUGCCUUAAUUAUUAAUACACAUAAGGCUAUGUAA ..(((((((......(((((.(((...((((((..((((((..-------.))))))......(((....)))))))))..)))...)))))..((((........)))))))))))... ( -34.00) >DroYak_CAF1 26388 117 + 1 CC---AGCCAAUCUCGAGGCCGUGAAAGAUCUUUUGAGAUCUGAGAACUGAGAUCUCUGACCAGCCAGACGGCAAGGUCGACACCAUGCCUUAAUUAUUAAUACACAUAAGGCUAUGUAA ..---((((......(((((.(((...((((((..(((((((.(....).)))))))......(((....)))))))))..)))...)))))..((((........))))))))...... ( -33.80) >consensus CU___AGCCAAUCUCGAGGCCGUGAAAGAUCUUUUGAGAUCUG_______AGAUCUCUGACCAGCCAGACGGCAAGGUCGACACCAUGCCUUAAUUAUUAAUACACAUAAGGCUAUGUAA .....((((......(((((.(((...((((((..(((((((........)))))))......(((....)))))))))..)))...)))))..((((........))))))))...... (-32.24 = -31.76 + -0.48)



| Location | 15,975,832 – 15,975,942 |
|---|---|
| Length | 110 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.16 |
| Mean single sequence MFE | -36.60 |
| Consensus MFE | -36.06 |
| Energy contribution | -35.78 |
| Covariance contribution | -0.28 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.72 |
| Structure conservation index | 0.99 |
| SVM decision value | 5.45 |
| SVM RNA-class probability | 0.999987 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15975832 110 - 22224390 UUACAUAGCCUUAUGUGUAUUAAUAAUUAAGGCAUGGUUUCGACCUUGCCAUCUGGCUGGUCAGAGAUCU-------CAGAUCUCAAAAGAUCUUUAACGGCCUCGAGAUUGGCU---GG .......((((((..(((....)))..))))))..((((((((((..(((....))).)))).)))))).-------.((((((....))))))....(((((........))))---). ( -37.60) >DroSec_CAF1 43643 110 - 1 UUACAUAGCCUUAUGUGUAUUAAUAAUUAAGGCAUGGUGUCGACCUUGCCGUCUGGCUGGUCAGAGAUCU-------CAGAUCUCAAAAGAUCUUUCACGGCCUCGAGAUUGGCU---AG .((((((......))))))...........((((.(((....))).))))..(((((..(((.(((..(.-------.((((((....)))))).....)..)))..)))..)))---)) ( -35.60) >DroSim_CAF1 34779 110 - 1 UUACAUAGCCUUAUGUGUAUUAAUAAUUAAGGCAUGGUGUCGACCUUGCCGUCUGGCUGGUCAGAGAUCU-------CAGAUCUCAAAAGAUCUUUCACGGCCUCGAGAUUGGCU---AU ...(((.((((((..(((....)))..)))))))))(((..((((..(((....))).)))).((((((.-------..))))))...........)))((((........))))---.. ( -34.70) >DroEre_CAF1 42513 113 - 1 UUACAUAGCCUUAUGUGUAUUAAUAAUUAAGGCAUAGUGUCGACCUUGCCGGGUGGCUGGUCAGAGAUCU-------CAGAUCUCAAAAGAUCUUUCACGGCCUCGAGAUUGGCCACGAG .((((((......))))))...........((((..((....))..))))..(((((..(((.(((..(.-------.((((((....)))))).....)..)))..)))..)))))... ( -36.60) >DroYak_CAF1 26388 117 - 1 UUACAUAGCCUUAUGUGUAUUAAUAAUUAAGGCAUGGUGUCGACCUUGCCGUCUGGCUGGUCAGAGAUCUCAGUUCUCAGAUCUCAAAAGAUCUUUCACGGCCUCGAGAUUGGCU---GG ...(((.((((((..(((....)))..)))))))))..(((((((..(((....))).)))).(((((((........)))))))....)))......(((((........))))---). ( -38.50) >consensus UUACAUAGCCUUAUGUGUAUUAAUAAUUAAGGCAUGGUGUCGACCUUGCCGUCUGGCUGGUCAGAGAUCU_______CAGAUCUCAAAAGAUCUUUCACGGCCUCGAGAUUGGCU___AG ...(((.((((((..(((....)))..)))))))))(((..((((..(((....))).)))).(((((((........)))))))...........)))((((........))))..... (-36.06 = -35.78 + -0.28)

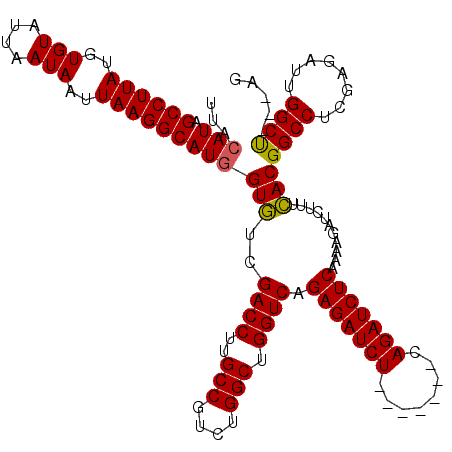
| Location | 15,975,869 – 15,975,981 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.17 |
| Mean single sequence MFE | -26.28 |
| Consensus MFE | -25.30 |
| Energy contribution | -25.50 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.24 |
| Structure conservation index | 0.96 |
| SVM decision value | 0.66 |
| SVM RNA-class probability | 0.813380 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15975869 112 - 22224390 UUGUUCG-UUUUUUUCCUUUCCCGCAUUUCGAUUAAGCCAUUACAUAGCCUUAUGUGUAUUAAUAAUUAAGGCAUGGUUUCGACCUUGCCAUCUGGCUGGUCAGAGAUCU-------CAG .......-...............(.((((((((((.((((.((((((......))))))...........((((.(((....))).))))...))))))))).)))))).-------... ( -26.10) >DroSec_CAF1 43680 108 - 1 ---UUCG-UUU-UUUCCUUUCCCGCAUUUCGAUUAAGCCAUUACAUAGCCUUAUGUGUAUUAAUAAUUAAGGCAUGGUGUCGACCUUGCCGUCUGGCUGGUCAGAGAUCU-------CAG ---....-...-...........(.((((((((((.((((.((((((......))))))...........((((.(((....))).))))...))))))))).)))))).-------... ( -26.30) >DroSim_CAF1 34816 111 - 1 UCUUUCG-UUU-UUUCCUUUCCCGCAUUUCGAUUAAGCCAUUACAUAGCCUUAUGUGUAUUAAUAAUUAAGGCAUGGUGUCGACCUUGCCGUCUGGCUGGUCAGAGAUCU-------CAG .......-...-...........(.((((((((((.((((.((((((......))))))...........((((.(((....))).))))...))))))))).)))))).-------... ( -26.30) >DroEre_CAF1 42553 105 - 1 G-------UUU-UUUCCUUUCCCGCAUUUCGAUUAAGCCAUUACAUAGCCUUAUGUGUAUUAAUAAUUAAGGCAUAGUGUCGACCUUGCCGGGUGGCUGGUCAGAGAUCU-------CAG .-------...-...........(.((((((((((.(((((((((((......))))))...........((((..((....))..))))..)))))))))).)))))).-------... ( -24.50) >DroYak_CAF1 26425 119 - 1 UUUUUGUUUUU-UUUCCUUUCCCGCAUUUCGAUUAAGCCAUUACAUAGCCUUAUGUGUAUUAAUAAUUAAGGCAUGGUGUCGACCUUGCCGUCUGGCUGGUCAGAGAUCUCAGUUCUCAG ...........-..................(((((.((((.((((((......))))))...........((((.(((....))).))))...))))))))).((((.......)))).. ( -28.20) >consensus U__UUCG_UUU_UUUCCUUUCCCGCAUUUCGAUUAAGCCAUUACAUAGCCUUAUGUGUAUUAAUAAUUAAGGCAUGGUGUCGACCUUGCCGUCUGGCUGGUCAGAGAUCU_______CAG .......................(.((((((((((.((((.((((((......))))))...........((((.(((....))).))))...))))))))).))))))........... (-25.30 = -25.50 + 0.20)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:10:25 2006