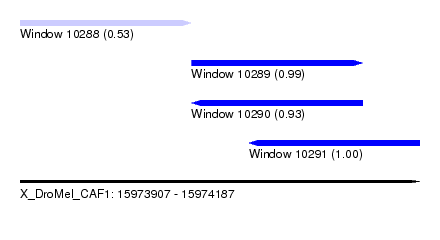
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 15,973,907 – 15,974,187 |
| Length | 280 |
| Max. P | 0.998038 |

| Location | 15,973,907 – 15,974,027 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 90.09 |
| Mean single sequence MFE | -22.63 |
| Consensus MFE | -17.49 |
| Energy contribution | -18.80 |
| Covariance contribution | 1.31 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.46 |
| Structure conservation index | 0.77 |
| SVM decision value | -0.02 |
| SVM RNA-class probability | 0.525912 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15973907 120 + 22224390 AAAUACAUAUAUUUUCUACAUUUCGGAAAUGCCUGAAAAGGUGAAUAUAGCUAUGUAUAUUUCACAUAAUAAUAUCAGGGCCAACAAUGUUAUUAGUGUGUGAAAAUAAAAGCUUAGUUU ..(((((((.((((((........))))))((((....)))).........))))))).(((((((((.(((((.((..(....)..)).))))).)))))))))............... ( -26.00) >DroSec_CAF1 41797 112 + 1 A----UAUAUAUUUUCUACAUUUCGGAAACGCCUGAAAAGGUGACUAUACCU----AUAAUUCACAUAAUAAUAUCAGGGCCAACAAUGUUAUUAGUGUGUGAAAAUAAAAGCUUGGUUU .----.......(((((.......)))))(((((....)))))((((.....----....((((((((.(((((.((..(....)..)).))))).))))))))..........)))).. ( -21.81) >DroSim_CAF1 32964 112 + 1 A----UAUAUAUUUUCUACAUUUCGGAAACGCCUGAAAGGGUGACUAUACCU----AUAAUUCACAUAAUAAUAUCAGGGCCAACAAUGUUAUUAGUGUGUGAAAAUAAAAGCUUGGUUU .----.......(((((.......)))))(((((....)))))((((.....----....((((((((.(((((.((..(....)..)).))))).))))))))..........)))).. ( -21.71) >DroEre_CAF1 40631 112 + 1 A----AAUGUAUUUUGUAUAUUUCGGAAACGCCUGAAAAGGUGACUAUACUU----AUAAUUCAUUUAAUAAUAUCAGGGCCAACAAUGUUAUUAGUGUGUGAAAAUAAAAGCAUAGUUU (----(((((((...)))))))).(....)((((....))))((((((.(((----.((.(((((((((((((((...(.....).))))))))))...)))))..)).))).)))))). ( -21.00) >consensus A____UAUAUAUUUUCUACAUUUCGGAAACGCCUGAAAAGGUGACUAUACCU____AUAAUUCACAUAAUAAUAUCAGGGCCAACAAUGUUAUUAGUGUGUGAAAAUAAAAGCUUAGUUU .............................(((((....))))).................((((((((.(((((.((..(....)..)).))))).))))))))................ (-17.49 = -18.80 + 1.31)



| Location | 15,974,027 – 15,974,147 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 98.50 |
| Mean single sequence MFE | -28.46 |
| Consensus MFE | -27.06 |
| Energy contribution | -27.46 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.84 |
| Structure conservation index | 0.95 |
| SVM decision value | 2.08 |
| SVM RNA-class probability | 0.987558 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15974027 120 + 22224390 UACGGCAUUUGCGUUCAUGGAAACUGUUUGGCAUGCGUGUUAAUUUGAAUAUAAAAGCAUAUUAAAAACGCGCCGCAACGCUCUUAAUUUCUCAAGCCACCAAAUGCAUGAUUAAUUCAU ....(((((((.((....(....).....(((..(((((((......(((((......)))))...))))))).((...))..............))))))))))))((((.....)))) ( -24.70) >DroSec_CAF1 41909 120 + 1 UACGGCAUUUGCGUUCAUGGAAACUGUUUGGCAUGCGUGUUAAUUUGAAUAUAAAAGCAUAUUAAAAACGCGCUGCAACGCGCUUAAUUUCUCAAGCCAUCAAAUGCAUGAUUAAUUCAU ....(((((((((((...(....)......(((.(((((((......(((((......)))))...)))))))))))))))((((........))))....))))))((((.....)))) ( -29.40) >DroSim_CAF1 33076 120 + 1 UACGGCAUUUGCGUUCAUGGAAACUGUUUGGCAUGCGUGUUAAUUUGAAUAUAAAAGCAUAUUAAAAACGCGCUGCAACGCGCUUAAUUUCUCAAGCCAUCAAAUGCAUGAUUAAUUCAU ....(((((((((((...(....)......(((.(((((((......(((((......)))))...)))))))))))))))((((........))))....))))))((((.....)))) ( -29.40) >DroEre_CAF1 40743 120 + 1 UACGGCAUUUGCGUUCAUGGAAACUGUUUGGCAUGCGUGUUAAUUUGAAUAUAAAAGCAUAUUAAAAACGCGCUGCAACGCGCUUAAUUUCUCAAGCCACCAAAUGCAUGAUUAAUUCAU ....(((((((((((...(....)......(((.(((((((......(((((......)))))...)))))))))))))))((((........))))....))))))((((.....)))) ( -29.40) >DroYak_CAF1 24520 120 + 1 UACGGCAUUUGCGUUCAUGGAAACUGUUUGGCAUGCGUGUUAAUUUGAAUAUAAAAGCAUAUUAAAAACGCGCUGCAACGCGCUUAAUUUCUCAAGCCACAAAAUGCAUGAUUAAUUCAU ....(((((((((((...(....)......(((.(((((((......(((((......)))))...)))))))))))))))((((........))))....))))))((((.....)))) ( -29.40) >consensus UACGGCAUUUGCGUUCAUGGAAACUGUUUGGCAUGCGUGUUAAUUUGAAUAUAAAAGCAUAUUAAAAACGCGCUGCAACGCGCUUAAUUUCUCAAGCCACCAAAUGCAUGAUUAAUUCAU ....(((((((((((...(....)......(((.(((((((......(((((......)))))...)))))))))))))))((((........))))....))))))((((.....)))) (-27.06 = -27.46 + 0.40)


| Location | 15,974,027 – 15,974,147 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 98.50 |
| Mean single sequence MFE | -27.99 |
| Consensus MFE | -27.26 |
| Energy contribution | -27.30 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.02 |
| Mean z-score | -1.49 |
| Structure conservation index | 0.97 |
| SVM decision value | 1.19 |
| SVM RNA-class probability | 0.927863 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15974027 120 - 22224390 AUGAAUUAAUCAUGCAUUUGGUGGCUUGAGAAAUUAAGAGCGUUGCGGCGCGUUUUUAAUAUGCUUUUAUAUUCAAAUUAACACGCAUGCCAAACAGUUUCCAUGAACGCAAAUGCCGUA ((((.....))))(((((((.((((.......(((((((((((......)))))))))))((((....................))))))))....((((....)))).))))))).... ( -25.75) >DroSec_CAF1 41909 120 - 1 AUGAAUUAAUCAUGCAUUUGAUGGCUUGAGAAAUUAAGCGCGUUGCAGCGCGUUUUUAAUAUGCUUUUAUAUUCAAAUUAACACGCAUGCCAAACAGUUUCCAUGAACGCAAAUGCCGUA ((((.....))))((((((....((((((....))))))(((((((((((.((((((((((((....)))))).)))..))).))).)))....((.......))))))))))))).... ( -28.70) >DroSim_CAF1 33076 120 - 1 AUGAAUUAAUCAUGCAUUUGAUGGCUUGAGAAAUUAAGCGCGUUGCAGCGCGUUUUUAAUAUGCUUUUAUAUUCAAAUUAACACGCAUGCCAAACAGUUUCCAUGAACGCAAAUGCCGUA ((((.....))))((((((....((((((....))))))(((((((((((.((((((((((((....)))))).)))..))).))).)))....((.......))))))))))))).... ( -28.70) >DroEre_CAF1 40743 120 - 1 AUGAAUUAAUCAUGCAUUUGGUGGCUUGAGAAAUUAAGCGCGUUGCAGCGCGUUUUUAAUAUGCUUUUAUAUUCAAAUUAACACGCAUGCCAAACAGUUUCCAUGAACGCAAAUGCCGUA ((((.....))))((((((....((((((....))))))(((((((((((.((((((((((((....)))))).)))..))).))).)))....((.......))))))))))))).... ( -28.70) >DroYak_CAF1 24520 120 - 1 AUGAAUUAAUCAUGCAUUUUGUGGCUUGAGAAAUUAAGCGCGUUGCAGCGCGUUUUUAAUAUGCUUUUAUAUUCAAAUUAACACGCAUGCCAAACAGUUUCCAUGAACGCAAAUGCCGUA ((((.....))))((((((....((((((....))))))(((((((((((.((((((((((((....)))))).)))..))).))).)))....((.......))))))))))))).... ( -28.10) >consensus AUGAAUUAAUCAUGCAUUUGGUGGCUUGAGAAAUUAAGCGCGUUGCAGCGCGUUUUUAAUAUGCUUUUAUAUUCAAAUUAACACGCAUGCCAAACAGUUUCCAUGAACGCAAAUGCCGUA ((((.....))))((((((....((((((....))))))(((((((((((.((((((((((((....)))))).)))..))).))).)))....((.......))))))))))))).... (-27.26 = -27.30 + 0.04)

| Location | 15,974,067 – 15,974,187 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.00 |
| Mean single sequence MFE | -34.07 |
| Consensus MFE | -31.56 |
| Energy contribution | -31.44 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.13 |
| Structure conservation index | 0.93 |
| SVM decision value | 2.99 |
| SVM RNA-class probability | 0.998038 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15974067 120 - 22224390 ACCGGUUGACAAUUCUCUGGGAAAAUCUGGCUGCCGCGCAAUGAAUUAAUCAUGCAUUUGGUGGCUUGAGAAAUUAAGAGCGUUGCGGCGCGUUUUUAAUAUGCUUUUAUAUUCAAAUUA ....((((.((((.((((.......((.(((..(((.(((.(((.....))))))...)))..))).)).......)))).))))))))((((.......))))................ ( -30.64) >DroSec_CAF1 41949 120 - 1 ACCGGUUGACAAUUCUCUGGGAAAAUCUGGCUGCCGCGCAAUGAAUUAAUCAUGCAUUUGAUGGCUUGAGAAAUUAAGCGCGUUGCAGCGCGUUUUUAAUAUGCUUUUAUAUUCAAAUUA .((((..((....)).))))(((......(((((.((((.........((((......)))).((((((....)))))))))).)))))((((.......)))).......)))...... ( -35.00) >DroSim_CAF1 33116 120 - 1 ACCGGUUGACAAUUCUCUGGGAAAAUCUGGCUGCCGCGCAAUGAAUUAAUCAUGCAUUUGAUGGCUUGAGAAAUUAAGCGCGUUGCAGCGCGUUUUUAAUAUGCUUUUAUAUUCAAAUUA .((((..((....)).))))(((......(((((.((((.........((((......)))).((((((....)))))))))).)))))((((.......)))).......)))...... ( -35.00) >DroEre_CAF1 40783 120 - 1 CCCGGUUGACAAUUGUCUGGGAAAAUCUGGCUGCCGCGCAAUGAAUUAAUCAUGCAUUUGGUGGCUUGAGAAAUUAAGCGCGUUGCAGCGCGUUUUUAAUAUGCUUUUAUAUUCAAAUUA ((((...(((....)))))))........(((((.((((.........((((......)))).((((((....)))))))))).)))))((((.......))))................ ( -35.20) >DroYak_CAF1 24560 120 - 1 CCCGGUUGACAAUUCUCUGGGAAAAACUGGCUGUCGCGCAAUGAAUUAAUCAUGCAUUUUGUGGCUUGAGAAAUUAAGCGCGUUGCAGCGCGUUUUUAAUAUGCUUUUAUAUUCAAAUUA (((((..((....)).)))))((((((..(((((.((((.((((.....))))..........((((((....)))))))))).)))))..))))))....................... ( -34.50) >consensus ACCGGUUGACAAUUCUCUGGGAAAAUCUGGCUGCCGCGCAAUGAAUUAAUCAUGCAUUUGGUGGCUUGAGAAAUUAAGCGCGUUGCAGCGCGUUUUUAAUAUGCUUUUAUAUUCAAAUUA .((((...........))))(((......(((((.((((.((((.....))))..........((((((....)))))))))).)))))((((.......)))).......)))...... (-31.56 = -31.44 + -0.12)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:10:21 2006