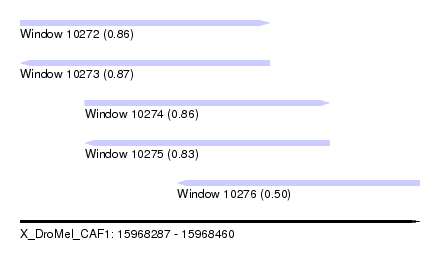
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 15,968,287 – 15,968,460 |
| Length | 173 |
| Max. P | 0.873900 |

| Location | 15,968,287 – 15,968,395 |
|---|---|
| Length | 108 |
| Sequences | 3 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 90.20 |
| Mean single sequence MFE | -41.47 |
| Consensus MFE | -30.90 |
| Energy contribution | -32.23 |
| Covariance contribution | 1.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.34 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.82 |
| SVM RNA-class probability | 0.858834 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15968287 108 + 22224390 UAUAGAUUUGCAUAUAAAGAUGGCU-----------AGAAGGAAAUAUCCGGGCUAGGAGUGAUUGGCUCCUGCCCAAGCGAAUGGCAGCUGAAAGGUUUCCUAGCGAGCAGCUGCUGA .....((((((..............-----------....(((....)))((((.((((((.....))))))))))..))))))((((((((....(((....)))...)))))))).. ( -40.10) >DroSec_CAF1 36329 119 + 1 UAUAGAUUUGCAUAUUAAGAUGGCUGGAUGUGACCAAGAAGGAAAUAUCCGGGCUAGGAGUGAUUGGCUCCAGCCCAAGCGAAUGACAGCUGAAAGGUUUCCUAGGGAGCAGCUGCUGA .....((((((..............((((((..((.....))..))))))(((((.(((((.....))))))))))..))))))..((((.(....(((((....)))))..).)))). ( -43.60) >DroSim_CAF1 27390 119 + 1 UAUAGAUUUGCAUAUUAAGAUGGCUGGAUGUGGCCAAGAAUGAAAUAUCCGGGCUAGGAGUGAUUGGCUCCUUCCCAAGCGAAUGGCAGCUGAAAGGUUUCCUAGGGAGCAGCUGCUGA .....((((((.........(((((......)))))..............(((..((((((.....)))))).)))..))))))(((((((.....(((((....)))))))))))).. ( -40.70) >consensus UAUAGAUUUGCAUAUUAAGAUGGCUGGAUGUG_CCAAGAAGGAAAUAUCCGGGCUAGGAGUGAUUGGCUCCUGCCCAAGCGAAUGGCAGCUGAAAGGUUUCCUAGGGAGCAGCUGCUGA .....((((((.............................(((....)))((((.((((((.....))))))))))..))))))((((((((..(((...)))......)))))))).. (-30.90 = -32.23 + 1.33)



| Location | 15,968,287 – 15,968,395 |
|---|---|
| Length | 108 |
| Sequences | 3 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 90.20 |
| Mean single sequence MFE | -31.83 |
| Consensus MFE | -25.25 |
| Energy contribution | -26.03 |
| Covariance contribution | 0.78 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.93 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.88 |
| SVM RNA-class probability | 0.873900 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15968287 108 - 22224390 UCAGCAGCUGCUCGCUAGGAAACCUUUCAGCUGCCAUUCGCUUGGGCAGGAGCCAAUCACUCCUAGCCCGGAUAUUUCCUUCU-----------AGCCAUCUUUAUAUGCAAAUCUAUA ...(((((((.......(....)....))))))).....((..(((((((((.......))))).))))(((....)))....-----------..............))......... ( -31.70) >DroSec_CAF1 36329 119 - 1 UCAGCAGCUGCUCCCUAGGAAACCUUUCAGCUGUCAUUCGCUUGGGCUGGAGCCAAUCACUCCUAGCCCGGAUAUUUCCUUCUUGGUCACAUCCAGCCAUCUUAAUAUGCAAAUCUAUA ...(((((((.......(....)....))))))).....((..(((((((((.......)))).)))))((((....((.....))....))))..............))......... ( -33.20) >DroSim_CAF1 27390 119 - 1 UCAGCAGCUGCUCCCUAGGAAACCUUUCAGCUGCCAUUCGCUUGGGAAGGAGCCAAUCACUCCUAGCCCGGAUAUUUCAUUCUUGGCCACAUCCAGCCAUCUUAAUAUGCAAAUCUAUA ...(((((((.......(....)....))))))).....(((((((.(((((.......)))))..)))))(((((.......((((........))))....)))))))......... ( -30.60) >consensus UCAGCAGCUGCUCCCUAGGAAACCUUUCAGCUGCCAUUCGCUUGGGCAGGAGCCAAUCACUCCUAGCCCGGAUAUUUCCUUCUUGG_CACAUCCAGCCAUCUUAAUAUGCAAAUCUAUA ...(((((((.......(....)....))))))).....((..(((((((((.......))))).))))(((....))).............................))......... (-25.25 = -26.03 + 0.78)


| Location | 15,968,315 – 15,968,421 |
|---|---|
| Length | 106 |
| Sequences | 3 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 88.22 |
| Mean single sequence MFE | -40.80 |
| Consensus MFE | -31.52 |
| Energy contribution | -33.30 |
| Covariance contribution | 1.78 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.74 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.81 |
| SVM RNA-class probability | 0.856089 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15968315 106 + 22224390 AGGAAAUAUCCGGGCUAGGAGUGAUUGGCUCCUGCCCAAGCGAAUGGCAGCUGAAAGGUUUCCUAGCGAGCAGCUGCUGAUCGAGAAUGGAUGCACCUUGAGUU----------GA (((...(((((((((.((((((.....))))))))))...(((.(((((((((....(((....)))...))))))))).))).....)))))..)))......----------.. ( -42.40) >DroSec_CAF1 36368 116 + 1 AGGAAAUAUCCGGGCUAGGAGUGAUUGGCUCCAGCCCAAGCGAAUGACAGCUGAAAGGUUUCCUAGGGAGCAGCUGCUGAACGAGGAUGGAUGCACCUUGAGCUUGUAGAAUGAAA .(((....)))(((((.(((((.....))))))))))((((......((((.(....(((((....)))))..).))))..(((((.((....))))))).))))........... ( -41.10) >DroSim_CAF1 27429 116 + 1 AUGAAAUAUCCGGGCUAGGAGUGAUUGGCUCCUUCCCAAGCGAAUGGCAGCUGAAAGGUUUCCUAGGGAGCAGCUGCUGAACGAGGAUGUAUGCACCUUGAGCUUGCAGAAUGAAA ...........(((..((((((.....)))))).)))((((....(((((((.....(((((....))))))))))))...(((((.((....))))))).))))........... ( -38.90) >consensus AGGAAAUAUCCGGGCUAGGAGUGAUUGGCUCCUGCCCAAGCGAAUGGCAGCUGAAAGGUUUCCUAGGGAGCAGCUGCUGAACGAGGAUGGAUGCACCUUGAGCUUG_AGAAUGAAA .(((....)))((((.((((((.....))))))))))((((....((((((((..(((...)))......))))))))...(((((.((....))))))).))))........... (-31.52 = -33.30 + 1.78)



| Location | 15,968,315 – 15,968,421 |
|---|---|
| Length | 106 |
| Sequences | 3 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 88.22 |
| Mean single sequence MFE | -32.77 |
| Consensus MFE | -26.31 |
| Energy contribution | -26.53 |
| Covariance contribution | 0.23 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.46 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.70 |
| SVM RNA-class probability | 0.826418 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15968315 106 - 22224390 UC----------AACUCAAGGUGCAUCCAUUCUCGAUCAGCAGCUGCUCGCUAGGAAACCUUUCAGCUGCCAUUCGCUUGGGCAGGAGCCAAUCACUCCUAGCCCGGAUAUUUCCU ..----------......(((...((((.....(((...(((((((.......(....)....)))))))...)))...(((((((((.......))))).))))))))....))) ( -36.00) >DroSec_CAF1 36368 116 - 1 UUUCAUUCUACAAGCUCAAGGUGCAUCCAUCCUCGUUCAGCAGCUGCUCCCUAGGAAACCUUUCAGCUGUCAUUCGCUUGGGCUGGAGCCAAUCACUCCUAGCCCGGAUAUUUCCU ...........((((....((.....))...........(((((((.......(....)....))))))).....))))(((((((((.......)))).)))))(((....))). ( -32.60) >DroSim_CAF1 27429 116 - 1 UUUCAUUCUGCAAGCUCAAGGUGCAUACAUCCUCGUUCAGCAGCUGCUCCCUAGGAAACCUUUCAGCUGCCAUUCGCUUGGGAAGGAGCCAAUCACUCCUAGCCCGGAUAUUUCAU ........((((..(....).))))...((((.((....(((((((.......(....)....)))))))....))...(((.(((((.......)))))..)))))))....... ( -29.70) >consensus UUUCAUUCU_CAAGCUCAAGGUGCAUCCAUCCUCGUUCAGCAGCUGCUCCCUAGGAAACCUUUCAGCUGCCAUUCGCUUGGGCAGGAGCCAAUCACUCCUAGCCCGGAUAUUUCCU ............................((((.((....(((((((.......(....)....)))))))....))...(((((((((.......))))).))))))))....... (-26.31 = -26.53 + 0.23)


| Location | 15,968,355 – 15,968,460 |
|---|---|
| Length | 105 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 84.10 |
| Mean single sequence MFE | -26.50 |
| Consensus MFE | -17.36 |
| Energy contribution | -17.44 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.68 |
| Structure conservation index | 0.66 |
| SVM decision value | -0.07 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15968355 105 - 22224390 CACGAUCUUUAGUCCGAUAA-AAGAGCUUUCCUCUCUAUCUC----------AACU----CAAGGUGCAUCCAUUCUCGAUCAGCAGCUGCUCGCUAGGAAACCUUUCAGCUGCCAUUCG ....(((((.(((..((((.-.((((.....)))).))))..----------.)))----.)))))...........(((...(((((((.......(....)....)))))))...))) ( -22.60) >DroSec_CAF1 36408 115 - 1 CACGAUCUUGAGUCCGAUAA-AAGAGCUUUCCUCUCUAUCUUUCAUUCUACAAGCU----CAAGGUGCAUCCAUCCUCGUUCAGCAGCUGCUCCCUAGGAAACCUUUCAGCUGUCAUUCG ....(((((((((..((((.-.((((.....)))).)))).............)))----))))))...........((....(((((((.......(....)....)))))))....)) ( -21.86) >DroSim_CAF1 27469 115 - 1 CACGAUCUUCAGUCCGAUAA-AAGAGCUUUCCUCUCUAUCUUUCAUUCUGCAAGCU----CAAGGUGCAUACAUCCUCGUUCAGCAGCUGCUCCCUAGGAAACCUUUCAGCUGCCAUUCG ..(((......(..(((...-..((((((......................)))))----)..((((....)))).)))..).(((((((.......(....)....)))))))...))) ( -22.95) >DroEre_CAF1 35295 115 - 1 CACGAUCUUGAGUUCGAUAA-AAGAGCUUUCCGCUCUAACUUUUAUUCCACUAGCU----GAAGAUGCAUCCAACCUUGGUCAGCAGCUGCUCCCUAGGAAACCUUUCAGCUGCCAUUCG ...((((..(.(((.(((..-.(((((.....)))))................((.----......))))).))))..)))).(((((((.......(....)....)))))))...... ( -28.20) >DroYak_CAF1 19197 116 - 1 CACGAUCUUGAGUCCGAUAAAAAGAGCUUUUCGCUCUAUCUUUUAUUCUACGGGCAUCUACGGGAUGCAUCCAUCCUCGAUCAGCAGCUGCCCCCUAGGAAACCUUUCAGCUGCCA---- ...((((....((((((((((((((((.....)))))...))))))....)))))......((((((....)))))).)))).(((((((.......(....)....)))))))..---- ( -36.90) >consensus CACGAUCUUGAGUCCGAUAA_AAGAGCUUUCCUCUCUAUCUUUCAUUCUACAAGCU____CAAGGUGCAUCCAUCCUCGAUCAGCAGCUGCUCCCUAGGAAACCUUUCAGCUGCCAUUCG ...((((...............((((.......))))..........................((((....))))...)))).(((((((.......(....)....)))))))...... (-17.36 = -17.44 + 0.08)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:10:07 2006