
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 15,963,284 – 15,963,423 |
| Length | 139 |
| Max. P | 0.997798 |

| Location | 15,963,284 – 15,963,394 |
|---|---|
| Length | 110 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 74.14 |
| Mean single sequence MFE | -20.58 |
| Consensus MFE | -13.62 |
| Energy contribution | -14.50 |
| Covariance contribution | 0.88 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.83 |
| Structure conservation index | 0.66 |
| SVM decision value | 1.39 |
| SVM RNA-class probability | 0.949022 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
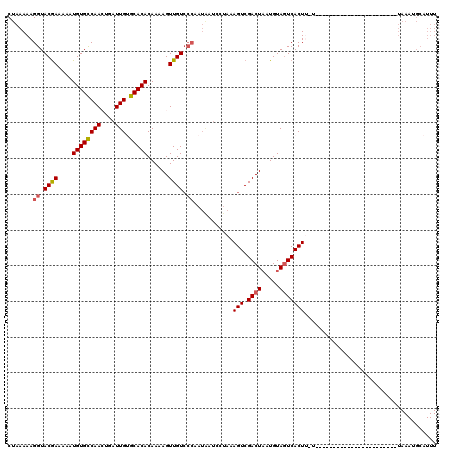
>X_DroMel_CAF1 15963284 110 + 22224390 CUAAAAAGGUACGAAAAAUGUGCCAACUGAUUGUGCACACAAAAGUUGUCCCAAUAAUCCGAAAGUCGACUAAUGUAGUCACUUUUCAUA---------CAUUUA-AAUUAAAUGCAUUU .......((.((((....((((((((....))).)))))......)))).))........((((((.(((((...)))))))))))....---------((((((-...))))))..... ( -20.40) >DroSec_CAF1 31433 94 + 1 CUAAAAAGGUACAAAAAAUGUGCCAACUGAUUGUGCACACAAAAGUUGUCCCAAUAAUCCUAAAGUCGACUAAUGUAGUCACU--------------------------UAAAUGCAUUU .......((.((((....((((((((....))).)))))......)))).))..........((((.(((((...))))))))--------------------------).......... ( -18.10) >DroSim_CAF1 22528 94 + 1 CUAAAAAGGUACGAAAAAUGUGCCAACUGAUUGUGCACACAAAAGUUGUCCCAAUAAUCCUAAAGUCGACUUAUGUAGUCACU--------------------------UAAAUGCAUUU .......((.((((....((((((((....))).)))))......)))).))..........((((.((((.....)))))))--------------------------).......... ( -17.90) >DroEre_CAF1 30064 108 + 1 CUAAAAAGGUACGAAAAAUGUGCCAAGUGAUUGAGCACACAAAAGUUGU------------AAAGUCGACUAAUGUACUCACUUCUCGGUUUUUUUGUGUGUUCAUAAUUAAAUGCAUUU .......((((((.....)))))).(((...(((((((((((((((((.------------.((((..((....))....))))..)))..))))))))))))))..))).......... ( -26.60) >DroYak_CAF1 13664 90 + 1 CUAAAAAGGUACGAAGAAUGUGUCAAGUGAUUGAGCACACAAAAGUUGUCCCAAUGAUCAUAUAGUCGACUAAUGUAGUCACUUUUUGGA------------------------------ (((((((((.((((....((((((((....)))).))))......)))).))..((((.((((((....)).)))).)))).))))))).------------------------------ ( -19.90) >consensus CUAAAAAGGUACGAAAAAUGUGCCAACUGAUUGUGCACACAAAAGUUGUCCCAAUAAUCCUAAAGUCGACUAAUGUAGUCACUU_U_______________________UAAAUGCAUUU .......((.((((....((((((((....))).)))))......)))).))...........(((.((((.....)))))))..................................... (-13.62 = -14.50 + 0.88)

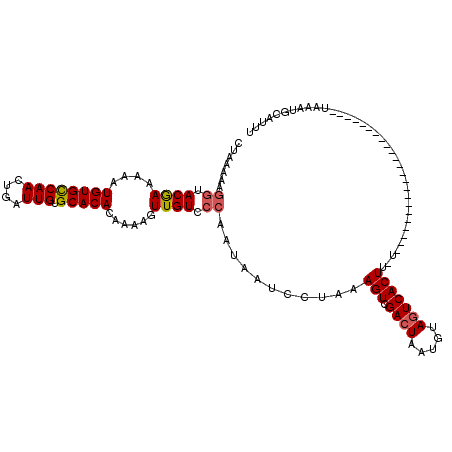
| Location | 15,963,324 – 15,963,423 |
|---|---|
| Length | 99 |
| Sequences | 3 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 78.34 |
| Mean single sequence MFE | -16.80 |
| Consensus MFE | -13.40 |
| Energy contribution | -13.40 |
| Covariance contribution | 0.01 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.58 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.89 |
| SVM RNA-class probability | 0.875429 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15963324 99 + 22224390 AAAAGUUGUCCCAAUAAUCCGAAAGUCGACUAAUGUAGUCACUUUUCAUACAUUUAAAUUAAAUGCAUUUUCAAUUGAAUUAUUGAAGCAGUUUAUUUU----------- ..((..(((..(((((((.(((((((.(((((...)))))))))))....((((((...))))))...........).)))))))..)))..)).....----------- ( -16.50) >DroSec_CAF1 31473 94 + 1 AAAAGUUGUCCCAAUAAUCCUAAAGUCGACUAAUGUAGUCACU----------------UAAAUGCAUUUUCGAUUGAAUCAUUGAAACAGUUGACUUUAAAAUGCCAAA ....................(((((((((((((((((......----------------....))))))((((((......))))))..))))))))))).......... ( -17.40) >DroSim_CAF1 22568 94 + 1 AAAAGUUGUCCCAAUAAUCCUAAAGUCGACUUAUGUAGUCACU----------------UAAAUGCAUUUUCGAUUGAAUCAUUGAAACAGUUGACUUUAAAAUGCCAAA ....................(((((((((((.(((((......----------------....)))))(((((((......))))))).))))))))))).......... ( -16.50) >consensus AAAAGUUGUCCCAAUAAUCCUAAAGUCGACUAAUGUAGUCACU________________UAAAUGCAUUUUCGAUUGAAUCAUUGAAACAGUUGACUUUAAAAUGCCAAA ....................(((((((((((.(((((..........................)))))(((((((......))))))).))))))))))).......... (-13.40 = -13.40 + 0.01)


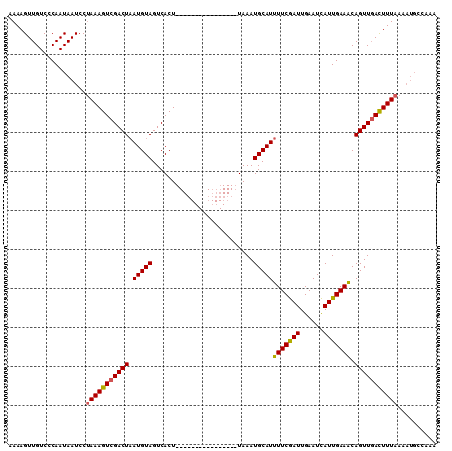
| Location | 15,963,324 – 15,963,423 |
|---|---|
| Length | 99 |
| Sequences | 3 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 78.34 |
| Mean single sequence MFE | -22.57 |
| Consensus MFE | -16.02 |
| Energy contribution | -16.13 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.74 |
| Structure conservation index | 0.71 |
| SVM decision value | 2.93 |
| SVM RNA-class probability | 0.997798 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15963324 99 - 22224390 -----------AAAAUAAACUGCUUCAAUAAUUCAAUUGAAAAUGCAUUUAAUUUAAAUGUAUGAAAAGUGACUACAUUAGUCGACUUUCGGAUUAUUGGGACAACUUUU -----------.........((..((((((((((........(((((((((...)))))))))((((..((((((...))))))..))))))))))))))..))...... ( -24.10) >DroSec_CAF1 31473 94 - 1 UUUGGCAUUUUAAAGUCAACUGUUUCAAUGAUUCAAUCGAAAAUGCAUUUA----------------AGUGACUACAUUAGUCGACUUUAGGAUUAUUGGGACAACUUUU .(((((........))))).(((..(((((((((....(......)....(----------------((((((((...))))).))))..)))))))))..)))...... ( -21.90) >DroSim_CAF1 22568 94 - 1 UUUGGCAUUUUAAAGUCAACUGUUUCAAUGAUUCAAUCGAAAAUGCAUUUA----------------AGUGACUACAUAAGUCGACUUUAGGAUUAUUGGGACAACUUUU .(((((........))))).(((..(((((((((....(......)....(----------------(((((((.....)))).))))..)))))))))..)))...... ( -21.70) >consensus UUUGGCAUUUUAAAGUCAACUGUUUCAAUGAUUCAAUCGAAAAUGCAUUUA________________AGUGACUACAUUAGUCGACUUUAGGAUUAUUGGGACAACUUUU ....................((((((((((((((....(......).....................(((((((.....)))).)))...))))))))))))))...... (-16.02 = -16.13 + 0.11)


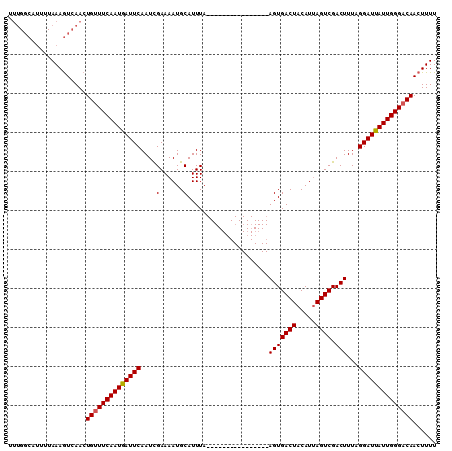
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:09:59 2006