
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 15,962,214 – 15,962,407 |
| Length | 193 |
| Max. P | 0.594049 |

| Location | 15,962,214 – 15,962,332 |
|---|---|
| Length | 118 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 89.03 |
| Mean single sequence MFE | -35.50 |
| Consensus MFE | -29.98 |
| Energy contribution | -31.48 |
| Covariance contribution | 1.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.42 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.10 |
| SVM RNA-class probability | 0.583705 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15962214 118 + 22224390 UUACUUAUCGGGUUUUUUUU--UGGGGCGUCACCAAGAUGAUGUCCAAACUGCGACUUCAAAUGCGGACCAUCCAGCCAAGUUGGCACAGGCUAUUCAAGCCAUCUCAUAGGCUUGGCCG .........((((.......--..((((((((......))))))))...(((((........)))))...)))).(((.....)))...(((....((((((........))))))))). ( -36.10) >DroSec_CAF1 30365 102 + 1 UUACUUAUCGGGUUU---UU--UGGGGCGUCACCAAGAUGAUGUCCA-------------AAUGCGGACCAUCCAGCCAAGUUGGCACAGGCUAUUCAAGCCAUCUCAUAGGCUUGGCCG ..........(((((---..--..((((((((......)))))))).-------------.....))))).....(((.....)))...(((....((((((........))))))))). ( -32.70) >DroEre_CAF1 29012 112 + 1 UUACUUAUCGGUUUU-------UGGGGCGUCACCAAGAUGAUGUCCAAACUGCGACUUGAAAUGCGGACCAUUCAGCCAAGUUGGCACAGGCUAUUCA-GCCAUCUCAUAGGCUUGGCCG .........(((...-------..((((((((......))))))))...(((((........)))))))).....(((.....)))...(((((...(-(((........))))))))). ( -35.00) >DroYak_CAF1 12585 120 + 1 UUACUUAUCGGUUUUUUUUUUGUGGGGCGUCACCAAGUUGAUGUCCAAACUGCGACUUGAAAUGCGGACCAUCCAGCCAAGUUGGCACAGGCUAUUCAAGCCAUCUCAUAGGCUUGGCCG ........((((......((((((((((((((......))))))))......(((((((....((((.....)).))))))))).)))))).....((((((........)))))))))) ( -38.20) >consensus UUACUUAUCGGGUUU___UU__UGGGGCGUCACCAAGAUGAUGUCCAAACUGCGACUUGAAAUGCGGACCAUCCAGCCAAGUUGGCACAGGCUAUUCAAGCCAUCUCAUAGGCUUGGCCG .........((.............((((((((......))))))))...(((((........))))).)).....(((.....)))...(((....((((((........))))))))). (-29.98 = -31.48 + 1.50)


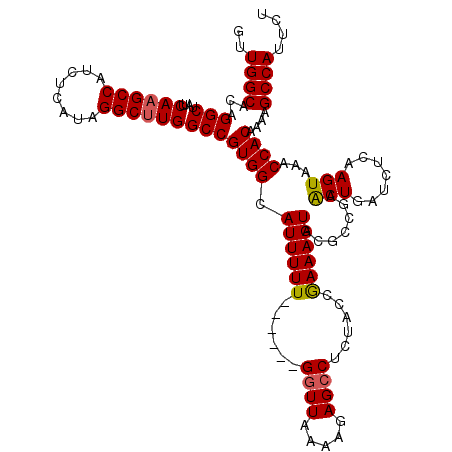
| Location | 15,962,292 – 15,962,407 |
|---|---|
| Length | 115 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.03 |
| Mean single sequence MFE | -31.87 |
| Consensus MFE | -27.25 |
| Energy contribution | -27.38 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.66 |
| Structure conservation index | 0.86 |
| SVM decision value | 0.12 |
| SVM RNA-class probability | 0.594049 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15962292 115 + 22224390 GUUGGCACAGGCUAUUCAAGCCAUCUCAUAGGCUUGGCCGUGGCAUUUUU-----GGUUUAAAAGAGCCUCUACCGAAAGUACGCCGAACUGAUCUCAAGUAAACCACAAAAGCCAUUCU ..((((...(((....((((((........)))))))))((((.((((((-----(((..............))))))))).......(((.......)))...))))....)))).... ( -31.74) >DroSec_CAF1 30427 120 + 1 GUUGGCACAGGCUAUUCAAGCCAUCUCAUAGGCUUGGCCGUGGCAUUUUUUUUUUUGGUUAAAAGAGCCUCUACCGAAAGUACGCCGAGCUGAUCUCAAGUAAACCACAAAAGCCAUUCU ..((((...((((.....))))........((((((((.(.(((.((((((.........))))))))).)...(....)...)))))))).....................)))).... ( -32.30) >DroEre_CAF1 29085 113 + 1 GUUGGCACAGGCUAUUCA-GCCAUCUCAUAGGCUUGGCCGUGGCAUUUUU------GGUUAAAAGAGCCUCUACCAAAAGUACGCCGAACUGAUCUCAAGUAAACCACAAAAGCCAUUCU ..((((...(((((...(-(((........)))))))))((((.((((((------(((.............))))))))).......(((.......)))...))))....)))).... ( -31.62) >DroYak_CAF1 12665 114 + 1 GUUGGCACAGGCUAUUCAAGCCAUCUCAUAGGCUUGGCCGUGGCAUUUUU------GGUUAAAAGAGCCUCUACCGAAAGUACGCCGAACUGAUCUCAAGUGAACCACAAAAGCCAUUCU ..((((...(((....((((((........)))))))))((((.((((((------(((.............))))))))).......(((.......)))...))))....)))).... ( -31.82) >consensus GUUGGCACAGGCUAUUCAAGCCAUCUCAUAGGCUUGGCCGUGGCAUUUUU______GGUUAAAAGAGCCUCUACCGAAAGUACGCCGAACUGAUCUCAAGUAAACCACAAAAGCCAUUCU ..((((...(((....((((((........)))))))))((((.((((((......((((.....))))......)))))).......(((.......)))...))))....)))).... (-27.25 = -27.38 + 0.12)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:09:56 2006