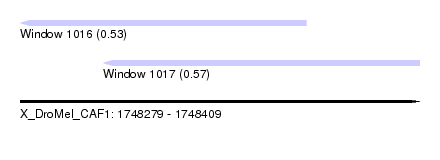
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 1,748,279 – 1,748,409 |
| Length | 130 |
| Max. P | 0.572791 |

| Location | 1,748,279 – 1,748,372 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 54.83 |
| Mean single sequence MFE | -17.08 |
| Consensus MFE | -5.57 |
| Energy contribution | -6.55 |
| Covariance contribution | 0.98 |
| Combinations/Pair | 1.67 |
| Mean z-score | -1.28 |
| Structure conservation index | 0.33 |
| SVM decision value | 0.00 |
| SVM RNA-class probability | 0.533908 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 1748279 93 - 22224390 CGCACCAUCUGCCAGUUGACGGCU-GGCGGAGAUUGUAAAUCCUCUCCACUAAUCACAAUAUUCCCUAUCAU------UCUUUGCCCAUCUCGUUUGUUC .(((......((((((.....)))-)))(((((..........)))))........................------....)))............... ( -18.30) >DroVir_CAF1 13964 93 - 1 ACUCUAUUGUAUUUAUAUAUCGCA-AUAACAUAAUUGAUAACAUGACAGAUUUUGGCUGUGCCUGCGAUUAA------ACUCUAUCAAAUUUCUCAUUCC ......(((.........((((((-....((....))........((((.......))))...))))))...------.......)))............ ( -9.41) >DroGri_CAF1 13815 93 - 1 AUCCUUUAAUA-----AUAUCGUACAUAACAUUGUUGCC-CCAAAAAAUGUUGUGGC-GAGCAUGCGAAUAAUUGUUGCUUACUUUUGACUUCUCAUUCU ...........-----...((((.(((((((((.((...-...)).)))))))))))-))(((.((((....)))))))..................... ( -16.10) >DroSec_CAF1 11531 93 - 1 CGCAUCAUCUGCCAGUUGACGGCU-GGCGGAGAUUGCAAAUCCUCUCCACUAAUCACAAUAUAUCCCAUCAU------UCUUUGCCCAUCUCGUUUGUUC .(((...(((((((((.....)))-))))))...)))...................................------...................... ( -20.20) >DroSim_CAF1 11758 93 - 1 CGCAUCAUCUGCCAGUUGACGGCU-GGCGGAGAUUGUAAAUCCUCUCCACUAAUCACAAUAUUCCCCAUCAU------UCUUUGCCCAUCUCGUUUGUUC .(((......((((((.....)))-)))(((((..........)))))........................------....)))............... ( -18.40) >DroEre_CAF1 11443 93 - 1 CACACCAUCUGCCAGUUGAUGGCU-GGCGGAGAUUGUGAAUCCUCUUUACUAAUCACAAUAUUUCCUAUCAU------UCUUUGCCAAUCUCGUUUGUUC ....(((((........))))).(-((((((((((((((..............)))))).............------)))))))))............. ( -20.05) >consensus CGCAUCAUCUGCCAGUUGACGGCU_GGCGGAGAUUGUAAAUCCUCUCCACUAAUCACAAUAUUUCCCAUCAU______UCUUUGCCCAUCUCGUUUGUUC .(((...(((((((((.....))).))))))...)))............................................................... ( -5.57 = -6.55 + 0.98)


| Location | 1,748,306 – 1,748,409 |
|---|---|
| Length | 103 |
| Sequences | 5 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 65.47 |
| Mean single sequence MFE | -25.69 |
| Consensus MFE | -10.44 |
| Energy contribution | -11.04 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.38 |
| Mean z-score | -1.75 |
| Structure conservation index | 0.41 |
| SVM decision value | 0.08 |
| SVM RNA-class probability | 0.572791 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 1748306 103 - 22224390 GCAGCAGUGAAUCCACUUUUGCAGGACUUCAAGGUGUCGCACCAUCUGCCAGUUGACGGCU-GGCGGAGAUUGUAAAUCCUCUCCACUAAUCACAAUAUUCCCU ((((.((((....)))).))))((((.(((((((((...)))).(((((((((.....)))-))))))..))).)).))))....................... ( -31.60) >DroGri_CAF1 13848 97 - 1 CCAAUUAAUAUUGAAUAUCUUUUAUACAUAUAUAAUUAUCCUUUAAUA-----AUAUCGUACAUAACAUUGUUGCC-CCAAAAAAUGUUGUGGC-GAGCAUGCG ........(((((((......(((((....)))))......)))))))-----...((((.(((((((((.((...-...)).)))))))))))-))....... ( -15.30) >DroSec_CAF1 11558 103 - 1 GCAGCAAUAAUUCCACUUUUGCAGGACUUCAAGGUGUCGCAUCAUCUGCCAGUUGACGGCU-GGCGGAGAUUGCAAAUCCUCUCCACUAAUCACAAUAUAUCCC ...((((...........)))).(((.....(((....(((...(((((((((.....)))-))))))...)))....))).)))................... ( -27.00) >DroSim_CAF1 11785 103 - 1 GCAGCAAUAAUUCCACUUUUGCAGGACUUCAAGGUGUCGCAUCAUCUGCCAGUUGACGGCU-GGCGGAGAUUGUAAAUCCUCUCCACUAAUCACAAUAUUCCCC ...((((...........)))).(((.....(((....(((...(((((((((.....)))-))))))...)))....))).)))................... ( -25.40) >DroEre_CAF1 11470 103 - 1 GCAGCAAUGAAUCCACUUUUGCAGGACUUCAAGGUGUCACACCAUCUGCCAGUUGAUGGCU-GGCGGAGAUUGUGAAUCCUCUUUACUAAUCACAAUAUUUCCU ...((((...........))))((((......((((...)))).(((((((((.....)))-)))))).(((((((..............)))))))...)))) ( -29.14) >consensus GCAGCAAUAAUUCCACUUUUGCAGGACUUCAAGGUGUCGCAUCAUCUGCCAGUUGACGGCU_GGCGGAGAUUGUAAAUCCUCUCCACUAAUCACAAUAUUUCCC ...(((((...(((.((...((..((((...(((((......)))))...))))....))..)).))).))))).............................. (-10.44 = -11.04 + 0.60)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:49:49 2006