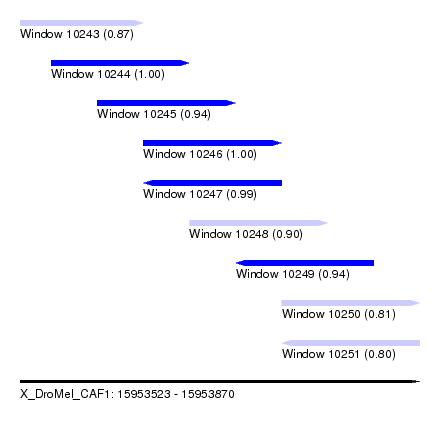
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 15,953,523 – 15,953,870 |
| Length | 347 |
| Max. P | 0.999161 |

| Location | 15,953,523 – 15,953,630 |
|---|---|
| Length | 107 |
| Sequences | 3 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 98.13 |
| Mean single sequence MFE | -40.86 |
| Consensus MFE | -40.83 |
| Energy contribution | -40.83 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.86 |
| Structure conservation index | 1.00 |
| SVM decision value | 0.86 |
| SVM RNA-class probability | 0.869766 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15953523 107 + 22224390 CAGCACCGCCUGCCGAUGAGCCCACCAUCGCCGAGUGAUGGAAGCCGCUCUUGUUGUUAUUCAAAUUGAGACUCAUUAGCUCCAGGAGCGGAUCCGUUGCGGGCAUU .......((((((.((((......(((((((...)))))))...(((((((((..((((.................))))..)))))))))...))))))))))... ( -40.83) >DroSec_CAF1 21879 107 + 1 CAGCGCCGCCUGCCGAUGAGCCUACCAUCGCCGAGUGAUGGAAGCCGCUCUUGUUGUUAUUCAAAUUGAGACUCAUUAGCUCCAGGAGCGGAUCCGUUGCGGGCAUC .......((((((.((((...((.(((((((...))))))).))(((((((((..((((.................))))..)))))))))...))))))))))... ( -40.93) >DroSim_CAF1 16400 107 + 1 CAGCGCCGCCUGCCGAUGAGCCCACCAUCGCCGAGUGAUGGAAGCCGCUCUUGUUGUUAUUCAAAUUGAGACUCAUUAGCUCCAGGAGCGGAUCCGUUGCGGGCAUC .......((((((.((((......(((((((...)))))))...(((((((((..((((.................))))..)))))))))...))))))))))... ( -40.83) >consensus CAGCGCCGCCUGCCGAUGAGCCCACCAUCGCCGAGUGAUGGAAGCCGCUCUUGUUGUUAUUCAAAUUGAGACUCAUUAGCUCCAGGAGCGGAUCCGUUGCGGGCAUC .......((((((.((((......(((((((...)))))))...(((((((((..((((.................))))..)))))))))...))))))))))... (-40.83 = -40.83 + -0.00)


| Location | 15,953,550 – 15,953,670 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 98.00 |
| Mean single sequence MFE | -46.39 |
| Consensus MFE | -43.47 |
| Energy contribution | -44.81 |
| Covariance contribution | 1.34 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.43 |
| Structure conservation index | 0.94 |
| SVM decision value | 3.41 |
| SVM RNA-class probability | 0.999161 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15953550 120 + 22224390 UCGCCGAGUGAUGGAAGCCGCUCUUGUUGUUAUUCAAAUUGAGACUCAUUAGCUCCAGGAGCGGAUCCGUUGCGGGCAUUACGGGUGCAUGGAGUAAACCAUGAUCAUCGCUGGUCGACA (((((.((((((((...(((((((((..((((.................))))..)))))))))((((((((....))..)))))).(((((......))))).)))))))))).))).. ( -44.83) >DroSec_CAF1 21906 120 + 1 UCGCCGAGUGAUGGAAGCCGCUCUUGUUGUUAUUCAAAUUGAGACUCAUUAGCUCCAGGAGCGGAUCCGUUGCGGGCAUCACGGGUGCAUGGAGUAAUCCAUGAUCAUCGCUGGUCGACA ..(((..(..(((((..(((((((((..((((.................))))..))))))))).)))))..).))).((((.((((((((((....)))))).....)))).)).)).. ( -48.83) >DroSim_CAF1 16427 120 + 1 UCGCCGAGUGAUGGAAGCCGCUCUUGUUGUUAUUCAAAUUGAGACUCAUUAGCUCCAGGAGCGGAUCCGUUGCGGGCAUCACGGGUGCAUGGAGUAAUCCAUGAUCAUCGCUGGUCGACA ..(((..(..(((((..(((((((((..((((.................))))..))))))))).)))))..).))).((((.((((((((((....)))))).....)))).)).)).. ( -48.83) >DroEre_CAF1 20529 120 + 1 UCGCCGAGUGAUGGAAGCCGCUCUUGUUAUUAUUCAAAUUGAGACUCAUUAGCUCCAGGAGCGGAUCCGUUGCGGGCAUCACGGGUGCAUGGAGUAGUCCAUGAUCGUCGCUGGUCGACA ..(((..(..(((((..(((((((((..............(((.((....)))))))))))))).)))))..).)))......(((.((((((....)))))))))((((.....)))). ( -44.33) >DroYak_CAF1 3972 120 + 1 UCGCCGAGUGAUGGAAGCCGCUCUUGUUGUUAUUCAAAUUGAGACUCAUUAGCUCCAGGAGCGGAUCGGUUGCGGGCAUCACGGGUGCAUGGAGUAAUCCAUGAUCAUCGCUGGUCGACA ..(((..(..((.((..(((((((((..((((.................))))..))))))))).)).))..).))).((((.((((((((((....)))))).....)))).)).)).. ( -45.13) >consensus UCGCCGAGUGAUGGAAGCCGCUCUUGUUGUUAUUCAAAUUGAGACUCAUUAGCUCCAGGAGCGGAUCCGUUGCGGGCAUCACGGGUGCAUGGAGUAAUCCAUGAUCAUCGCUGGUCGACA ..(((..(..(((((..(((((((((..((((.................))))..))))))))).)))))..).))).((((.((((((((((....)))))).....)))).)).)).. (-43.47 = -44.81 + 1.34)



| Location | 15,953,590 – 15,953,710 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 97.33 |
| Mean single sequence MFE | -43.36 |
| Consensus MFE | -39.18 |
| Energy contribution | -40.16 |
| Covariance contribution | 0.98 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.62 |
| Structure conservation index | 0.90 |
| SVM decision value | 1.25 |
| SVM RNA-class probability | 0.935671 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15953590 120 + 22224390 GAGACUCAUUAGCUCCAGGAGCGGAUCCGUUGCGGGCAUUACGGGUGCAUGGAGUAAACCAUGAUCAUCGCUGGUCGACAACAGUUUGCGAUGCAAAUUGAGAUUCGAACUGUGACGAUC ...........(((((((.((((....)))).)..((((.....)))).)))))).......((((.((((...((((...((((((((...))))))))....))))...)))).)))) ( -37.90) >DroSec_CAF1 21946 120 + 1 GAGACUCAUUAGCUCCAGGAGCGGAUCCGUUGCGGGCAUCACGGGUGCAUGGAGUAAUCCAUGAUCAUCGCUGGUCGACAGCAGCUUGCGAUGCAAAUUGAGAUUCGAACUGUGACGAUC ...........((((...)))).((((.((..((((((((.(((((.((((((....))))))......((((.....)))).))))).)))))...(((.....))).)))..)))))) ( -44.40) >DroSim_CAF1 16467 120 + 1 GAGACUCAUUAGCUCCAGGAGCGGAUCCGUUGCGGGCAUCACGGGUGCAUGGAGUAAUCCAUGAUCAUCGCUGGUCGACAGCAGCUUGCGAUGCAAAUUGAGAUUCGAACUGUGACGAUC ...........((((...)))).((((.((..((((((((.(((((.((((((....))))))......((((.....)))).))))).)))))...(((.....))).)))..)))))) ( -44.40) >DroEre_CAF1 20569 120 + 1 GAGACUCAUUAGCUCCAGGAGCGGAUCCGUUGCGGGCAUCACGGGUGCAUGGAGUAGUCCAUGAUCGUCGCUGGUCGACAGCAGCUUGCGAUGCAAAUUGAGAUUCGAGCUGUGACGAUC (.((((.....(((((((.((((....)))).)..((((.....)))).)))))))))))..(((((((((.(.((((...(((.((((...)))).)))....)))).).))))))))) ( -45.70) >DroYak_CAF1 4012 120 + 1 GAGACUCAUUAGCUCCAGGAGCGGAUCGGUUGCGGGCAUCACGGGUGCAUGGAGUAAUCCAUGAUCAUCGCUGGUCGACAGCAGCUUGCGAUGCAAAUUGAGAUUCGAACUGUGACGAUC ...........((((...)))).(((((.(..((((((((.(((((.((((((....))))))......((((.....)))).))))).)))))...(((.....))).)))..)))))) ( -44.40) >consensus GAGACUCAUUAGCUCCAGGAGCGGAUCCGUUGCGGGCAUCACGGGUGCAUGGAGUAAUCCAUGAUCAUCGCUGGUCGACAGCAGCUUGCGAUGCAAAUUGAGAUUCGAACUGUGACGAUC ...........((((...)))).((((.((..((((((((.(((((.((((((....))))))......((((.....)))).))))).)))))...(((.....))).)))..)))))) (-39.18 = -40.16 + 0.98)

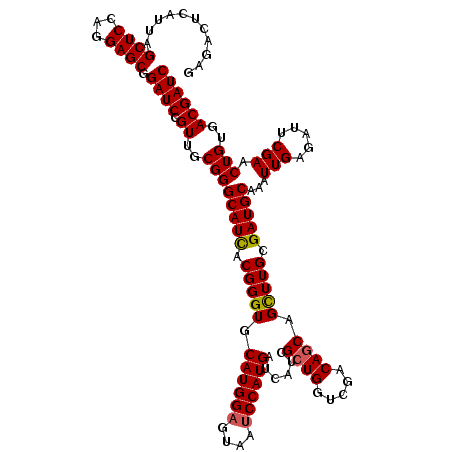
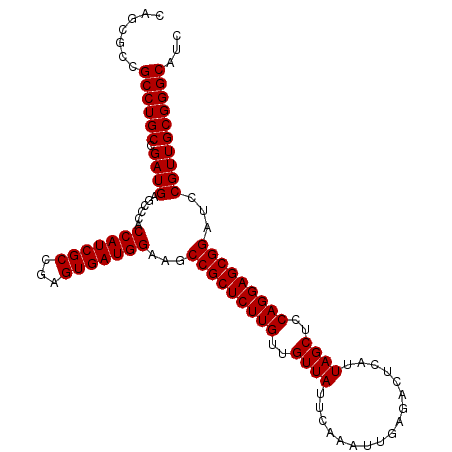
| Location | 15,953,630 – 15,953,750 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.83 |
| Mean single sequence MFE | -47.74 |
| Consensus MFE | -41.38 |
| Energy contribution | -42.52 |
| Covariance contribution | 1.14 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.61 |
| Structure conservation index | 0.87 |
| SVM decision value | 2.85 |
| SVM RNA-class probability | 0.997371 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15953630 120 + 22224390 ACGGGUGCAUGGAGUAAACCAUGAUCAUCGCUGGUCGACAACAGUUUGCGAUGCAAAUUGAGAUUCGAACUGUGACGAUCGCGGCUGCGUUUCGAGGGAAAUGCCGCAUUGCAGCCAUCU ..((.(((((((......))))((((.((((...((((...((((((((...))))))))....))))...)))).))))(((((...(((((....))))))))))...))).)).... ( -45.00) >DroSec_CAF1 21986 120 + 1 ACGGGUGCAUGGAGUAAUCCAUGAUCAUCGCUGGUCGACAGCAGCUUGCGAUGCAAAUUGAGAUUCGAACUGUGACGAUCGCGGCUGCGCUUCGAGGGAAAUGCCGCAUUGCAGCCAUCU ..((.((((((((....)))))((((.((((...((((...(((.((((...)))).)))....))))...)))).))))(((((..(.(.....).)....)))))...))).)).... ( -44.50) >DroSim_CAF1 16507 120 + 1 ACGGGUGCAUGGAGUAAUCCAUGAUCAUCGCUGGUCGACAGCAGCUUGCGAUGCAAAUUGAGAUUCGAACUGUGACGAUCGCGGCUGCGUUUCGAGGGAAAUGCCGCAUUGCAGCCAUCU ..((.((((((((....)))))((((.((((...((((...(((.((((...)))).)))....))))...)))).))))(((((...(((((....))))))))))...))).)).... ( -45.80) >DroEre_CAF1 20609 120 + 1 ACGGGUGCAUGGAGUAGUCCAUGAUCGUCGCUGGUCGACAGCAGCUUGCGAUGCAAAUUGAGAUUCGAGCUGUGACGAUCGCGGCUGCGUUUCGAGGGAAACGCGGCAUUGCAGCCAUCU ..((.((((((((....)))))(((((((((.(.((((...(((.((((...)))).)))....)))).).)))))))))...((((((((((....))))))))))...))).)).... ( -55.00) >DroYak_CAF1 4052 120 + 1 ACGGGUGCAUGGAGUAAUCCAUGAUCAUCGCUGGUCGACAGCAGCUUGCGAUGCAAAUUGAGAUUCGAACUGUGACGAUCGCGGCUGCGUUUCGAGGGAAAUGCGGCAUUGCAGCCAUCU ..((.((((((((....)))))((((.((((...((((...(((.((((...)))).)))....))))...)))).))))...((((((((((....))))))))))...))).)).... ( -48.40) >consensus ACGGGUGCAUGGAGUAAUCCAUGAUCAUCGCUGGUCGACAGCAGCUUGCGAUGCAAAUUGAGAUUCGAACUGUGACGAUCGCGGCUGCGUUUCGAGGGAAAUGCCGCAUUGCAGCCAUCU ..((.((((((((....)))))((((.((((.(.((((...(((.((((...)))).)))....)))).).)))).))))...((.(((((((....))))))).))...))).)).... (-41.38 = -42.52 + 1.14)



| Location | 15,953,630 – 15,953,750 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.83 |
| Mean single sequence MFE | -38.68 |
| Consensus MFE | -36.80 |
| Energy contribution | -36.72 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.76 |
| Structure conservation index | 0.95 |
| SVM decision value | 2.33 |
| SVM RNA-class probability | 0.992515 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15953630 120 - 22224390 AGAUGGCUGCAAUGCGGCAUUUCCCUCGAAACGCAGCCGCGAUCGUCACAGUUCGAAUCUCAAUUUGCAUCGCAAACUGUUGUCGACCAGCGAUGAUCAUGGUUUACUCCAUGCACCCGU ..((((.(((...(((((.((((....))))....)))))(((((((.(.(.((((....((.(((((...))))).))...)))).).).)))))))((((......))))))).)))) ( -38.80) >DroSec_CAF1 21986 120 - 1 AGAUGGCUGCAAUGCGGCAUUUCCCUCGAAGCGCAGCCGCGAUCGUCACAGUUCGAAUCUCAAUUUGCAUCGCAAGCUGCUGUCGACCAGCGAUGAUCAUGGAUUACUCCAUGCACCCGU ..((((.(((...(((((.((((....))))....)))))(((((((.(.(.((((....((.(((((...))))).))...)))).).).)))))))(((((....)))))))).)))) ( -38.50) >DroSim_CAF1 16507 120 - 1 AGAUGGCUGCAAUGCGGCAUUUCCCUCGAAACGCAGCCGCGAUCGUCACAGUUCGAAUCUCAAUUUGCAUCGCAAGCUGCUGUCGACCAGCGAUGAUCAUGGAUUACUCCAUGCACCCGU ..((((.(((...(((((.((((....))))....)))))(((((((.(.(.((((....((.(((((...))))).))...)))).).).)))))))(((((....)))))))).)))) ( -38.80) >DroEre_CAF1 20609 120 - 1 AGAUGGCUGCAAUGCCGCGUUUCCCUCGAAACGCAGCCGCGAUCGUCACAGCUCGAAUCUCAAUUUGCAUCGCAAGCUGCUGUCGACCAGCGACGAUCAUGGACUACUCCAUGCACCCGU ..((((.(((...((.(((((((....))))))).))...(((((((..(((((((..(.......)..)))..))))((((.....)))))))))))(((((....)))))))).)))) ( -42.60) >DroYak_CAF1 4052 120 - 1 AGAUGGCUGCAAUGCCGCAUUUCCCUCGAAACGCAGCCGCGAUCGUCACAGUUCGAAUCUCAAUUUGCAUCGCAAGCUGCUGUCGACCAGCGAUGAUCAUGGAUUACUCCAUGCACCCGU ..((((.(((...((.((.((((....)))).)).))...(((((((.(.(.((((....((.(((((...))))).))...)))).).).)))))))(((((....)))))))).)))) ( -34.70) >consensus AGAUGGCUGCAAUGCGGCAUUUCCCUCGAAACGCAGCCGCGAUCGUCACAGUUCGAAUCUCAAUUUGCAUCGCAAGCUGCUGUCGACCAGCGAUGAUCAUGGAUUACUCCAUGCACCCGU ..((((.(((...(((((.((((....))))....)))))(((((((.(.(.((((....((.(((((...))))).))...)))).).).)))))))((((......))))))).)))) (-36.80 = -36.72 + -0.08)


| Location | 15,953,670 – 15,953,790 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 97.83 |
| Mean single sequence MFE | -43.88 |
| Consensus MFE | -42.22 |
| Energy contribution | -42.10 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.40 |
| Structure conservation index | 0.96 |
| SVM decision value | 0.99 |
| SVM RNA-class probability | 0.896298 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15953670 120 + 22224390 ACAGUUUGCGAUGCAAAUUGAGAUUCGAACUGUGACGAUCGCGGCUGCGUUUCGAGGGAAAUGCCGCAUUGCAGCCAUCUAUAUUGCACUUGUGGUGCAGCUUGAGGUGGACGCUCUGGU .(((.(((((((((...............(((((.....)))))..(((((((....))))))).)))))))))((((((...((((((.....))))))....)))))).....))).. ( -42.40) >DroSec_CAF1 22026 120 + 1 GCAGCUUGCGAUGCAAAUUGAGAUUCGAACUGUGACGAUCGCGGCUGCGCUUCGAGGGAAAUGCCGCAUUGCAGCCAUCUAUAUUGCACUUGUGGUGCAGCUUGAGGUGGACGCUCUGGU ..(((.((((((((.......((..((..(((((.....)))))...))..))..((......))))))))))(((((((...((((((.....))))))....)))))).))))..... ( -40.30) >DroSim_CAF1 16547 120 + 1 GCAGCUUGCGAUGCAAAUUGAGAUUCGAACUGUGACGAUCGCGGCUGCGUUUCGAGGGAAAUGCCGCAUUGCAGCCAUCUAUAUUGCACUUGUGGUGCAGCUUGAGGUGGACGCUCUGGU ..(((.((((((((...............(((((.....)))))..(((((((....))))))).))))))))(((((((...((((((.....))))))....)))))).))))..... ( -42.50) >DroEre_CAF1 20649 120 + 1 GCAGCUUGCGAUGCAAAUUGAGAUUCGAGCUGUGACGAUCGCGGCUGCGUUUCGAGGGAAACGCGGCAUUGCAGCCAUCUAUAUUGCACUUGUGGUGCAGCUUGAGGUGGACGCUCUGGU ..(((.((((((((.............(((((((.....)))))))(((((((....))))))).))))))))(((((((...((((((.....))))))....)))))).))))..... ( -50.10) >DroYak_CAF1 4092 120 + 1 GCAGCUUGCGAUGCAAAUUGAGAUUCGAACUGUGACGAUCGCGGCUGCGUUUCGAGGGAAAUGCGGCAUUGCAGCCAUCUAUAUUGCACUUGUGGUGCAGCUUGAGGUGGACGCUCUGGU .(((...(((.(((((..............((((.....))))((((((((((....)))))))))).))))).((((((...((((((.....))))))....)))))).))).))).. ( -44.10) >consensus GCAGCUUGCGAUGCAAAUUGAGAUUCGAACUGUGACGAUCGCGGCUGCGUUUCGAGGGAAAUGCCGCAUUGCAGCCAUCUAUAUUGCACUUGUGGUGCAGCUUGAGGUGGACGCUCUGGU ..((((((((((((...............(((((.....)))))..(((((((....))))))).)))))))))((((((...((((((.....))))))....))))))..)))..... (-42.22 = -42.10 + -0.12)


| Location | 15,953,710 – 15,953,830 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 98.33 |
| Mean single sequence MFE | -31.33 |
| Consensus MFE | -29.47 |
| Energy contribution | -29.87 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.59 |
| Structure conservation index | 0.94 |
| SVM decision value | 1.30 |
| SVM RNA-class probability | 0.939374 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15953710 120 - 22224390 GGUGAGAUAUUCCAAAAUCACUUUCAUCUCAAGACGCACCACCAGAGCGUCCACCUCAAGCUGCACCACAAGUGCAAUAUAGAUGGCUGCAAUGCGGCAUUUCCCUCGAAACGCAGCCGC ((((.(((........))).............(((((.........)))))))))....(((((((.....)))))........((((((....(((........)))....)))))))) ( -31.80) >DroSec_CAF1 22066 120 - 1 GGUGAGAUAUUCCAAAACCACUUUCAUCUCAAGACGCACCACCAGAGCGUCCACCUCAAGCUGCACCACAAGUGCAAUAUAGAUGGCUGCAAUGCGGCAUUUCCCUCGAAGCGCAGCCGC ((((((((.................)))))..(((((.........))))).)))....(((((((.....)))))........((((((....(((........)))....)))))))) ( -31.43) >DroSim_CAF1 16587 120 - 1 GGUGAGAUAUUCCAAAACCACUUUCAUCUCAAGACGCACCACCAGAGCGUCCACCUCAAGCUGCACCACAAGUGCAAUAUAGAUGGCUGCAAUGCGGCAUUUCCCUCGAAACGCAGCCGC ((((((((.................)))))..(((((.........))))).)))....(((((((.....)))))........((((((....(((........)))....)))))))) ( -31.43) >DroEre_CAF1 20689 120 - 1 GGUGAGAUAUUCCAAAACCACUUUCAUCUCAAGACGCACCACCAGAGCGUCCACCUCAAGCUGCACCACAAGUGCAAUAUAGAUGGCUGCAAUGCCGCGUUUCCCUCGAAACGCAGCCGC ((((((((.................)))))..(((((.........))))).)))....(((((((.....)))).........(((......)))(((((((....))))))))))... ( -33.33) >DroYak_CAF1 4132 120 - 1 GGUGAGAUAUUCCAAAAUCACUUUCAUCUCAAGACGCACCACCAGAGCGUCCACCUCAAGCUGCACCACAAGUGCAAUAUAGAUGGCUGCAAUGCCGCAUUUCCCUCGAAACGCAGCCGC ((((.(((........))).............(((((.........)))))))))....(((((((.....)))))........((((((......................)))))))) ( -28.65) >consensus GGUGAGAUAUUCCAAAACCACUUUCAUCUCAAGACGCACCACCAGAGCGUCCACCUCAAGCUGCACCACAAGUGCAAUAUAGAUGGCUGCAAUGCGGCAUUUCCCUCGAAACGCAGCCGC ((((((((.................)))))..(((((.........))))).)))....(((((((.....)))))........((((((....(((........)))....)))))))) (-29.47 = -29.87 + 0.40)


| Location | 15,953,750 – 15,953,870 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 98.17 |
| Mean single sequence MFE | -41.46 |
| Consensus MFE | -39.14 |
| Energy contribution | -39.74 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.36 |
| Structure conservation index | 0.94 |
| SVM decision value | 0.64 |
| SVM RNA-class probability | 0.808111 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15953750 120 + 22224390 AUAUUGCACUUGUGGUGCAGCUUGAGGUGGACGCUCUGGUGGUGCGUCUUGAGAUGAAAGUGAUUUUGGAAUAUCUCACCGCAGGCGGUGCACUUUAGGGGGUUCUCCUUAUCGAUUGGC ...((((((.....))))))..(((((.((((.((((((.(((((.....(((((((((....)))).....)))))((((....))))))))))))))).)))).)))))......... ( -42.70) >DroSec_CAF1 22106 120 + 1 AUAUUGCACUUGUGGUGCAGCUUGAGGUGGACGCUCUGGUGGUGCGUCUUGAGAUGAAAGUGGUUUUGGAAUAUCUCACCGCAGGCGGUGCACUUUAGGGGGUUCUCCUUAUCGAUGGGC ...((((((.....))))))..(((((.((((.((((((.(((((.....((((((.(((....)))....))))))((((....))))))))))))))).)))).)))))......... ( -42.00) >DroSim_CAF1 16627 120 + 1 AUAUUGCACUUGUGGUGCAGCUUGAGGUGGACGCUCUGGUGGUGCGUCUUGAGAUGAAAGUGGUUUUGGAAUAUCUCACCGCAGGCGGUGCACUUUAGGGGGUUCUCCUUAUCGAUGGGC ...((((((.....))))))..(((((.((((.((((((.(((((.....((((((.(((....)))....))))))((((....))))))))))))))).)))).)))))......... ( -42.00) >DroEre_CAF1 20729 120 + 1 AUAUUGCACUUGUGGUGCAGCUUGAGGUGGACGCUCUGGUGGUGCGUCUUGAGAUGAAAGUGGUUUUGGAAUAUCUCACCGCAGGCGGUGCACUUUAGGGGGUUCUCCUUAUCGAUGGGC ...((((((.....))))))..(((((.((((.((((((.(((((.....((((((.(((....)))....))))))((((....))))))))))))))).)))).)))))......... ( -42.00) >DroYak_CAF1 4172 120 + 1 AUAUUGCACUUGUGGUGCAGCUUGAGGUGGACGCUCUGGUGGUGCGUCUUGAGAUGAAAGUGAUUUUGGAAUAUCUCACCACAGGCGGUGCACUUAAGGGGAUUCUCCUUAUCGAUGGGC ....(((((.....)))))(((((.(((((((((.(.....).)))))).(((((((((....)))).....)))))))).))))).((.((..(((((((...)))))))....)).)) ( -38.60) >consensus AUAUUGCACUUGUGGUGCAGCUUGAGGUGGACGCUCUGGUGGUGCGUCUUGAGAUGAAAGUGGUUUUGGAAUAUCUCACCGCAGGCGGUGCACUUUAGGGGGUUCUCCUUAUCGAUGGGC ...((((((.....))))))..(((((.((((.((((((.(((((.....((((((.(((....)))....))))))((((....))))))))))))))).)))).)))))......... (-39.14 = -39.74 + 0.60)



| Location | 15,953,750 – 15,953,870 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 98.17 |
| Mean single sequence MFE | -26.80 |
| Consensus MFE | -25.47 |
| Energy contribution | -25.47 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.34 |
| Structure conservation index | 0.95 |
| SVM decision value | 0.60 |
| SVM RNA-class probability | 0.796605 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15953750 120 - 22224390 GCCAAUCGAUAAGGAGAACCCCCUAAAGUGCACCGCCUGCGGUGAGAUAUUCCAAAAUCACUUUCAUCUCAAGACGCACCACCAGAGCGUCCACCUCAAGCUGCACCACAAGUGCAAUAU (((........(((.......)))...(((((..((.((.((((.(((........))).............(((((.........))))))))).)).))))))).....).))..... ( -26.50) >DroSec_CAF1 22106 120 - 1 GCCCAUCGAUAAGGAGAACCCCCUAAAGUGCACCGCCUGCGGUGAGAUAUUCCAAAACCACUUUCAUCUCAAGACGCACCACCAGAGCGUCCACCUCAAGCUGCACCACAAGUGCAAUAU ((.(.......(((.......)))...(((((..((.((.((((((((.................)))))..(((((.........))))).))).)).))))))).....).))..... ( -26.13) >DroSim_CAF1 16627 120 - 1 GCCCAUCGAUAAGGAGAACCCCCUAAAGUGCACCGCCUGCGGUGAGAUAUUCCAAAACCACUUUCAUCUCAAGACGCACCACCAGAGCGUCCACCUCAAGCUGCACCACAAGUGCAAUAU ((.(.......(((.......)))...(((((..((.((.((((((((.................)))))..(((((.........))))).))).)).))))))).....).))..... ( -26.13) >DroEre_CAF1 20729 120 - 1 GCCCAUCGAUAAGGAGAACCCCCUAAAGUGCACCGCCUGCGGUGAGAUAUUCCAAAACCACUUUCAUCUCAAGACGCACCACCAGAGCGUCCACCUCAAGCUGCACCACAAGUGCAAUAU ((.(.......(((.......)))...(((((..((.((.((((((((.................)))))..(((((.........))))).))).)).))))))).....).))..... ( -26.13) >DroYak_CAF1 4172 120 - 1 GCCCAUCGAUAAGGAGAAUCCCCUUAAGUGCACCGCCUGUGGUGAGAUAUUCCAAAAUCACUUUCAUCUCAAGACGCACCACCAGAGCGUCCACCUCAAGCUGCACCACAAGUGCAAUAU ((.(.....(((((.......))))).(((((..((.((.((((.(((........))).............(((((.........))))))))).)).))))))).....).))..... ( -29.10) >consensus GCCCAUCGAUAAGGAGAACCCCCUAAAGUGCACCGCCUGCGGUGAGAUAUUCCAAAACCACUUUCAUCUCAAGACGCACCACCAGAGCGUCCACCUCAAGCUGCACCACAAGUGCAAUAU ((.........(((.......)))...(((((..((.((.((((((((.................)))))..(((((.........))))).))).)).))))))).......))..... (-25.47 = -25.47 + 0.00)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:09:45 2006