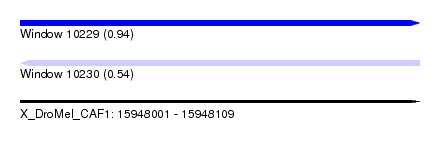
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 15,948,001 – 15,948,109 |
| Length | 108 |
| Max. P | 0.937045 |

| Location | 15,948,001 – 15,948,109 |
|---|---|
| Length | 108 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 74.89 |
| Mean single sequence MFE | -29.12 |
| Consensus MFE | -18.19 |
| Energy contribution | -20.75 |
| Covariance contribution | 2.56 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.98 |
| Structure conservation index | 0.62 |
| SVM decision value | 1.26 |
| SVM RNA-class probability | 0.937045 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15948001 108 + 22224390 ----------CGAUGCUAUGUAAUACUAUAUAAC--ACUGUGACGAGAUGGUCCCUGAGAUUUAAAUUCAGAUUUGAAUCUGAAAGUUACUCGCACAUUUGUGAUAUACAGCCAUCUUCG ----------...((.((((((....)))))).)--)......(((((((((...(.((((((((((....)))))))))).)..((...(((((....)))))...)).)))))).))) ( -24.40) >DroSec_CAF1 16421 115 + 1 UUUA--GUAUCUAUGCUGUGUAGUACUAUCAUACGGACAGGGGC--GAUGGUCCCUGAGAUUUAAAUUCAGAUUCGAAUCUGA-AGUAGCUCGCACAUUUGUGAUAUACAGCCAUCUUCG ....--........((((((((.........(((...(((((((--....))))))).........(((((((....))))))-))))..(((((....)))))))))))))........ ( -33.60) >DroSim_CAF1 10924 116 + 1 UUAAAUGUAUCUAUGCUGUGUAAUACUAUAUAAC--ACAGGGGC--GAUGGUCCCUGAGAUUUAAAUUCAGAUUCGAAUCUGAAAGUAGCUCGCACAUUUGUGAUAUACAGCCAUCUUCG ..............((((((((.((((.......--.(((((((--....))))))).........(((((((....)))))))))))..(((((....)))))))))))))........ ( -34.30) >DroEre_CAF1 15523 91 + 1 -------UAUCUAUGCUGCGUAUUAA----------------------AUCUGCCUGAGAUUUAAACUCGGAUUCGAAUCUGAGAGUAGCCCGCACAUUGGUGAUGUACAGCCAGCUUCG -------.......(((((...((((----------------------((((.....)))))))).(((((((....))))))).))))).......(((((........)))))..... ( -24.20) >consensus _______UAUCUAUGCUGUGUAAUACUAUAUAAC__ACAGGGGC__GAUGGUCCCUGAGAUUUAAAUUCAGAUUCGAAUCUGAAAGUAGCUCGCACAUUUGUGAUAUACAGCCAUCUUCG ..............((((((((...............(((((((......))))))).((.(((..(((((((....)))))))..))).)).(((....))).))))))))........ (-18.19 = -20.75 + 2.56)

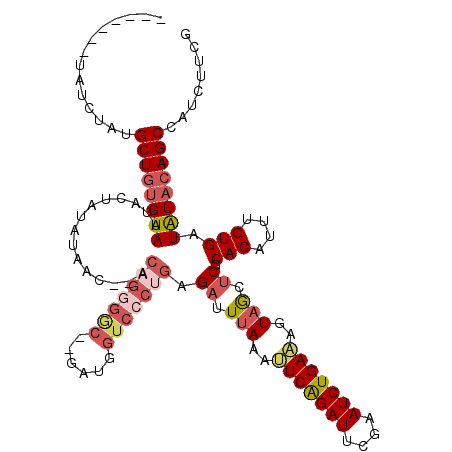

| Location | 15,948,001 – 15,948,109 |
|---|---|
| Length | 108 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 74.89 |
| Mean single sequence MFE | -26.05 |
| Consensus MFE | -15.66 |
| Energy contribution | -15.35 |
| Covariance contribution | -0.31 |
| Combinations/Pair | 1.22 |
| Mean z-score | -1.63 |
| Structure conservation index | 0.60 |
| SVM decision value | 0.01 |
| SVM RNA-class probability | 0.539995 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15948001 108 - 22224390 CGAAGAUGGCUGUAUAUCACAAAUGUGCGAGUAACUUUCAGAUUCAAAUCUGAAUUUAAAUCUCAGGGACCAUCUCGUCACAGU--GUUAUAUAGUAUUACAUAGCAUCG---------- ....((((((((((((.(((...((((((((.....(((((((....))))))).......))).(((.....)))).))))))--).)))))))).........)))).---------- ( -27.10) >DroSec_CAF1 16421 115 - 1 CGAAGAUGGCUGUAUAUCACAAAUGUGCGAGCUACU-UCAGAUUCGAAUCUGAAUUUAAAUCUCAGGGACCAUC--GCCCCUGUCCGUAUGAUAGUACUACACAGCAUAGAUAC--UAAA .((((.(((((((((((.....)))))).)))))))-))(((((..(((....)))..)))))(((((......--..))))).........((((((((.......))).)))--)).. ( -28.70) >DroSim_CAF1 10924 116 - 1 CGAAGAUGGCUGUAUAUCACAAAUGUGCGAGCUACUUUCAGAUUCGAAUCUGAAUUUAAAUCUCAGGGACCAUC--GCCCCUGU--GUUAUAUAGUAUUACACAGCAUAGAUACAUUUAA ....(((((((((((((.....))))))(((.((..(((((((....)))))))..))...)))..)).)))))--....((((--((.(((...))).))))))............... ( -26.30) >DroEre_CAF1 15523 91 - 1 CGAAGCUGGCUGUACAUCACCAAUGUGCGGGCUACUCUCAGAUUCGAAUCCGAGUUUAAAUCUCAGGCAGAU----------------------UUAAUACGCAGCAUAGAUA------- ....((((.((((((((.....))))))))..........((((((....))))))(((((((.....))))----------------------))).....)))).......------- ( -22.10) >consensus CGAAGAUGGCUGUAUAUCACAAAUGUGCGAGCUACUUUCAGAUUCGAAUCUGAAUUUAAAUCUCAGGGACCAUC__GCCCCUGU__GUUAUAUAGUAUUACACAGCAUAGAUA_______ ........(((((....(((....))).(((.....(((((((....))))))).......))).....................................))))).............. (-15.66 = -15.35 + -0.31)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:09:23 2006