
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 15,937,930 – 15,938,090 |
| Length | 160 |
| Max. P | 0.606318 |

| Location | 15,937,930 – 15,938,050 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.64 |
| Mean single sequence MFE | -30.57 |
| Consensus MFE | -27.93 |
| Energy contribution | -28.60 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.74 |
| Structure conservation index | 0.91 |
| SVM decision value | 0.09 |
| SVM RNA-class probability | 0.578408 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
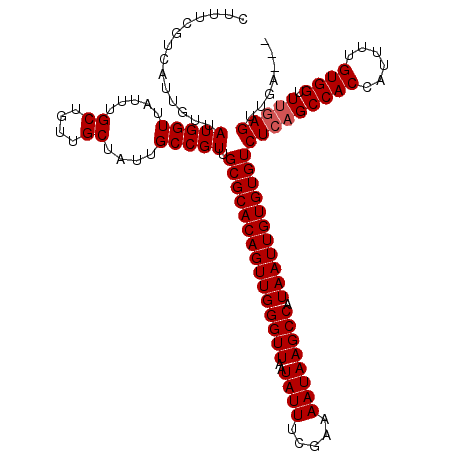
>X_DroMel_CAF1 15937930 120 + 22224390 CUUUCGUCAUUGUUAUGGUUAUUUGCUGUUGCUAUUGCCGUUGCGCACAGUUGGGUUAAUAUUUCGAAAAUAAGCCAUAAUUGUGUGUCUCAGCCACCAUUUUGUGGUUUUAGUUGAGUU ..............(((((.....((....))....))))).(((((((((((((((..((((.....)))))))).)))))))))))((((((.(((((...)))))....)))))).. ( -30.60) >DroSec_CAF1 6202 117 + 1 CUUUCGUCAUUGUUAUGGUUAUUUGCUGUUGCUAUUGCCGUUGCGCACAGUUGGGUUAAUAUUUCGAAAAUAAGCCGUAAUUGUGUGUCUCAGCCACCAUUUUGUGGUUUGAGUUGA--- ..............(((((.....((....))....))))).(((((((((((((((..((((.....)))))))).)))))))))))(((((((((......)))).)))))....--- ( -31.00) >DroEre_CAF1 5935 117 + 1 CUUUCGUCAUUGUUAUGGUUAUUUGCUGUUGCUAUUGCCGUUGCGCACAGUUGGGUUAAUAUUUCGAAAAUAAGCCAUAAUUGUGUGUCUCUGCCACCAUUUUGUGGUUUGAGUUGA--- ..............(((((.....((....))....))))).(((((((((((((((..((((.....)))))))).)))))))))))(((.(((((......)))))..)))....--- ( -30.10) >consensus CUUUCGUCAUUGUUAUGGUUAUUUGCUGUUGCUAUUGCCGUUGCGCACAGUUGGGUUAAUAUUUCGAAAAUAAGCCAUAAUUGUGUGUCUCAGCCACCAUUUUGUGGUUUGAGUUGA___ ..............(((((.....((....))....))))).(((((((((((((((..((((.....)))))))).)))))))))))(((((((((......)))).)))))....... (-27.93 = -28.60 + 0.67)


| Location | 15,937,970 – 15,938,090 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.63 |
| Mean single sequence MFE | -27.67 |
| Consensus MFE | -22.53 |
| Energy contribution | -23.20 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.07 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.03 |
| SVM RNA-class probability | 0.546818 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15937970 120 + 22224390 UUGCGCACAGUUGGGUUAAUAUUUCGAAAAUAAGCCAUAAUUGUGUGUCUCAGCCACCAUUUUGUGGUUUUAGUUGAGUUUGAGUUUGUGCUUGCUUUGGUUAUUUCCUUCAUUUUUUUU ..(((((((((((((((..((((.....)))))))).)))))))))))((((((.(((((...)))))....))))))...((((....)))).....((......))............ ( -26.70) >DroSec_CAF1 6242 113 + 1 UUGCGCACAGUUGGGUUAAUAUUUCGAAAAUAAGCCGUAAUUGUGUGUCUCAGCCACCAUUUUGUGGUUUGAGUUGA------GCUUGUGCUUGCUUUGGUUAUUUCCUUCAGUUUUUU- ..(((((((((((((((..((((.....)))))))).)))))))))))(((((((((......)))).)))))..((------((....))))(((..((......))...))).....- ( -30.10) >DroEre_CAF1 5975 113 + 1 UUGCGCACAGUUGGGUUAAUAUUUCGAAAAUAAGCCAUAAUUGUGUGUCUCUGCCACCAUUUUGUGGUUUGAGUUGA------GUUUGUGCUUGCUUUGGUUAUUUCCUUCUUUUUUUU- ..(((((((((((((((..((((.....)))))))).)))))))))))(((.(((((......)))))..)))..((------((....)))).....((......))...........- ( -26.20) >consensus UUGCGCACAGUUGGGUUAAUAUUUCGAAAAUAAGCCAUAAUUGUGUGUCUCAGCCACCAUUUUGUGGUUUGAGUUGA______GUUUGUGCUUGCUUUGGUUAUUUCCUUCAUUUUUUU_ ..(((((((((((((((..((((.....)))))))).)))))))))))(((((((((......)))).))))).........................((......))............ (-22.53 = -23.20 + 0.67)


| Location | 15,937,970 – 15,938,090 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.63 |
| Mean single sequence MFE | -18.13 |
| Consensus MFE | -15.93 |
| Energy contribution | -16.27 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.84 |
| Structure conservation index | 0.88 |
| SVM decision value | 0.15 |
| SVM RNA-class probability | 0.606318 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15937970 120 - 22224390 AAAAAAAAUGAAGGAAAUAACCAAAGCAAGCACAAACUCAAACUCAACUAAAACCACAAAAUGGUGGCUGAGACACACAAUUAUGGCUUAUUUUCGAAAUAUUAACCCAACUGUGCGCAA ............((......))...((..(((((........((((.((...((((.....)))))).))))...........(((.(((............))).)))..))))))).. ( -17.60) >DroSec_CAF1 6242 113 - 1 -AAAAAACUGAAGGAAAUAACCAAAGCAAGCACAAGC------UCAACUCAAACCACAAAAUGGUGGCUGAGACACACAAUUACGGCUUAUUUUCGAAAUAUUAACCCAACUGUGCGCAA -...........((((((((((......(((....))------)...((((..((((......)))).))))............)).))))))))......................... ( -18.90) >DroEre_CAF1 5975 113 - 1 -AAAAAAAAGAAGGAAAUAACCAAAGCAAGCACAAAC------UCAACUCAAACCACAAAAUGGUGGCAGAGACACACAAUUAUGGCUUAUUUUCGAAAUAUUAACCCAACUGUGCGCAA -...........((......))...((..(((((...------....(((...((((......))))..)))...........(((.(((............))).)))..))))))).. ( -17.90) >consensus _AAAAAAAUGAAGGAAAUAACCAAAGCAAGCACAAAC______UCAACUCAAACCACAAAAUGGUGGCUGAGACACACAAUUAUGGCUUAUUUUCGAAAUAUUAACCCAACUGUGCGCAA ............((......))...((..(((((.............(((...((((......))))..)))............((.(((............))).))...))))))).. (-15.93 = -16.27 + 0.33)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:09:15 2006