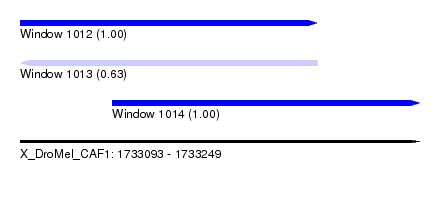
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 1,733,093 – 1,733,249 |
| Length | 156 |
| Max. P | 0.999935 |

| Location | 1,733,093 – 1,733,209 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 87.86 |
| Mean single sequence MFE | -57.72 |
| Consensus MFE | -52.58 |
| Energy contribution | -52.26 |
| Covariance contribution | -0.32 |
| Combinations/Pair | 1.12 |
| Mean z-score | -3.56 |
| Structure conservation index | 0.91 |
| SVM decision value | 4.66 |
| SVM RNA-class probability | 0.999935 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 1733093 116 + 22224390 UGAGGCAAUCC----GUACAGAGAGCAGGACUUACCAUGGUGAGAUACGGGUGUCGGAGGAGUAACAUUACCUGCUCCUGGCGCUCGUACUCGUGGGUCAGGGUGUCCAUUGCUCGCUGA ...((....))----...(((.(((((((((..(((.(((((((.((((((((((..(((((((........)))))))))))))))))))))....))).)))))))..))))).))). ( -47.40) >DroSec_CAF1 4312 118 + 1 UGGGGCAAUCC--GCGUACAGCGAGCAGGACUUACCAGGGUGAGAUACGGGUGUCGAAGGAGUAGCAUUACCUGCUCCUGGCGCUCGUACUCGCGGGUGAGGGUGUCCAUUGCUCGCUGG .(((....)))--.....((((((((((((((((((...(((((.((((((((((..((((((((......))))))))))))))))))))))).)))))....))))..))))))))). ( -62.00) >DroSim_CAF1 4866 118 + 1 UGGGGCAAUCC--GCGUACAGCGAGCAGGACUUACCAGGGUGAGAUACGGGUGUCGAAGGAGUAGCAUUACCUGCUCCUGGCGCUCGUACUCGCGGGUGAGGGUGUCCAUUGCUCGCUGG .(((....)))--.....((((((((((((((((((...(((((.((((((((((..((((((((......))))))))))))))))))))))).)))))....))))..))))))))). ( -62.00) >DroEre_CAF1 4407 120 + 1 UCGGACAAUCCGCGGGUAUAGCGAGCAAGACUUACCAGGGUGAUAUACGGGUGCCGGAGGAGCAGCAUCACCUGCUCCUGGCGCUCGUACUCGCGGGUCAGGGUGUCCAUUGCUCGCUGG .(((.....)))......((((((((((....((((..(((((..((((((((((..((((((((......)))))))))))))))))).))))....)..))))....)))))))))). ( -56.70) >DroYak_CAF1 4495 119 + 1 CCGUGUGGUU-ACGCGUAUAGCGAGCAAGACUUACCAGGGUAAGGUACGGGUGCCGAAGGAGCAGCAUGAUCUGCUCCUGGCGCUCGUACUCGCGGGUCAGGGUGUCCAUUGCUCGCUGG .(((((....-)))))..(((((((((((((((((....)))))(((((((((((..((((((((......)))))))))))))))))))))...((.(.....).)).)))))))))). ( -60.50) >consensus UGGGGCAAUCC__GCGUACAGCGAGCAGGACUUACCAGGGUGAGAUACGGGUGUCGAAGGAGUAGCAUUACCUGCUCCUGGCGCUCGUACUCGCGGGUCAGGGUGUCCAUUGCUCGCUGG ..................((((((((((..((.(((...(((((.((((((((((..((((((((......))))))))))))))))))))))).))).))((...)).)))))))))). (-52.58 = -52.26 + -0.32)

| Location | 1,733,093 – 1,733,209 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 87.86 |
| Mean single sequence MFE | -41.50 |
| Consensus MFE | -32.12 |
| Energy contribution | -32.72 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.58 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.19 |
| SVM RNA-class probability | 0.625546 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 1733093 116 - 22224390 UCAGCGAGCAAUGGACACCCUGACCCACGAGUACGAGCGCCAGGAGCAGGUAAUGUUACUCCUCCGACACCCGUAUCUCACCAUGGUAAGUCCUGCUCUCUGUAC----GGAUUGCCUCA .(((.(((((..((((............(((((((.(.(...((((..((((....)))).))))..).).)))).)))((....))..))))))))).)))...----((....))... ( -32.10) >DroSec_CAF1 4312 118 - 1 CCAGCGAGCAAUGGACACCCUCACCCGCGAGUACGAGCGCCAGGAGCAGGUAAUGCUACUCCUUCGACACCCGUAUCUCACCCUGGUAAGUCCUGCUCGCUGUACGC--GGAUUGCCCCA .(((((((((..((((...(((...(....)...))).((((((((((.....)))).......((.....))........))))))..))))))))))))).....--((......)). ( -39.80) >DroSim_CAF1 4866 118 - 1 CCAGCGAGCAAUGGACACCCUCACCCGCGAGUACGAGCGCCAGGAGCAGGUAAUGCUACUCCUUCGACACCCGUAUCUCACCCUGGUAAGUCCUGCUCGCUGUACGC--GGAUUGCCCCA .(((((((((..((((...(((...(....)...))).((((((((((.....)))).......((.....))........))))))..))))))))))))).....--((......)). ( -39.80) >DroEre_CAF1 4407 120 - 1 CCAGCGAGCAAUGGACACCCUGACCCGCGAGUACGAGCGCCAGGAGCAGGUGAUGCUGCUCCUCCGGCACCCGUAUAUCACCCUGGUAAGUCUUGCUCGCUAUACCCGCGGAUUGUCCGA ..((((((((..((((......(((.(.(((((((.(.(((((((((((......))))))))..))).).))))).)).)...)))..)))))))))))).......(((.....))). ( -47.60) >DroYak_CAF1 4495 119 - 1 CCAGCGAGCAAUGGACACCCUGACCCGCGAGUACGAGCGCCAGGAGCAGAUCAUGCUGCUCCUUCGGCACCCGUACCUUACCCUGGUAAGUCUUGCUCGCUAUACGCGU-AACCACACGG ..((((((((..((((..............(((((.(.(((((((((((......))))))))..))).).))))).((((....))))))))))))))))...((.((-....)).)). ( -48.20) >consensus CCAGCGAGCAAUGGACACCCUGACCCGCGAGUACGAGCGCCAGGAGCAGGUAAUGCUACUCCUUCGACACCCGUAUCUCACCCUGGUAAGUCCUGCUCGCUGUACGC__GGAUUGCCCCA .(((((((((..((((..........((........))((((((((((.....)))).......((.....))........))))))..))))))))))))).................. (-32.12 = -32.72 + 0.60)

| Location | 1,733,129 – 1,733,249 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.58 |
| Mean single sequence MFE | -54.59 |
| Consensus MFE | -52.18 |
| Energy contribution | -51.54 |
| Covariance contribution | -0.64 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.16 |
| Structure conservation index | 0.96 |
| SVM decision value | 3.93 |
| SVM RNA-class probability | 0.999709 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 1733129 120 + 22224390 UGAGAUACGGGUGUCGGAGGAGUAACAUUACCUGCUCCUGGCGCUCGUACUCGUGGGUCAGGGUGUCCAUUGCUCGCUGACUGACCGGUUUCACCAGGCGCCGCUUGCCCCGGAAUAUGU .(((.((((((((((..(((((((........))))))))))))))))))))(.((((.((((((((...((...((((......))))..))...)))))).)).)))))......... ( -47.20) >DroSec_CAF1 4350 120 + 1 UGAGAUACGGGUGUCGAAGGAGUAGCAUUACCUGCUCCUGGCGCUCGUACUCGCGGGUGAGGGUGUCCAUUGCUCGCUGGCUGACCGGCUUCACCAGGCGCCGCUUGCCCCGGAAUAUGU .(((.((((((((((..((((((((......)))))))))))))))))))))(.(((..((((((((...((...(((((....)))))..))...)))))).))..))))......... ( -57.10) >DroSim_CAF1 4904 120 + 1 UGAGAUACGGGUGUCGAAGGAGUAGCAUUACCUGCUCCUGGCGCUCGUACUCGCGGGUGAGGGUGUCCAUUGCUCGCUGGCUGACCGGUUUCACCAGGCGCCGCUUGCCCCGGAAUAUGU .(((.((((((((((..((((((((......)))))))))))))))))))))(.(((..((((((((...((...(((((....)))))..))...)))))).))..))))......... ( -55.50) >DroEre_CAF1 4447 120 + 1 UGAUAUACGGGUGCCGGAGGAGCAGCAUCACCUGCUCCUGGCGCUCGUACUCGCGGGUCAGGGUGUCCAUUGCUCGCUGGCUCACCGGCUUUACCAGGCGCCGCUUGCCUCGGAAUAUGU .....((((((((((..((((((((......)))))))))))))))))).((.(((((.((((((((........(((((....))))).......)))))).)).))).))))...... ( -53.26) >DroYak_CAF1 4534 120 + 1 UAAGGUACGGGUGCCGAAGGAGCAGCAUGAUCUGCUCCUGGCGCUCGUACUCGCGGGUCAGGGUGUCCAUUGCUCGCUGGCUGACCGGCUUCACCAGGCGCCGCUUGCCCCGGAAUAUGU ...((((((((((((..((((((((......))))))))))))))))))))((.((((.((((((((...((...(((((....)))))..))...)))))).)).))))))........ ( -59.90) >consensus UGAGAUACGGGUGUCGAAGGAGUAGCAUUACCUGCUCCUGGCGCUCGUACUCGCGGGUCAGGGUGUCCAUUGCUCGCUGGCUGACCGGCUUCACCAGGCGCCGCUUGCCCCGGAAUAUGU .....((((((((((..((((((((......)))))))))))))))))).(((.(((..((((((((...((...(((((....)))))..))...)))))).))..))))))....... (-52.18 = -51.54 + -0.64)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:49:46 2006