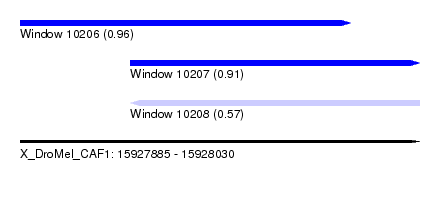
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 15,927,885 – 15,928,030 |
| Length | 145 |
| Max. P | 0.964000 |

| Location | 15,927,885 – 15,928,005 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 90.25 |
| Mean single sequence MFE | -30.34 |
| Consensus MFE | -25.86 |
| Energy contribution | -27.02 |
| Covariance contribution | 1.16 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.86 |
| Structure conservation index | 0.85 |
| SVM decision value | 1.56 |
| SVM RNA-class probability | 0.964000 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15927885 120 + 22224390 CUAGCACACUCUGUCCUUAAGUAUAUCUUGGGAUUAAAGUAUAUAUGUAUAUGUGACCUCUGUUGCGAAAUUUCGAUCCCUUCUUCCCUACGGGAUGAAGAUAGAGGUCAUCGUCUUGAU (.((.(((((..(((((.(((.....))))))))...)))............(((((((((((..(((....)))....((((.((((...)))).))))))))))))))).)))).).. ( -32.80) >DroSec_CAF1 26436 104 + 1 CUAGCACACUUUGUCCUUAAGUAUAUCUUGGGAUUAAAGUAUAUAUGUAUAUGUGACCUCUGUUGCGAAAUUUCGAUCC----------------UGAAGAUAGAGGUCAUCGUCUUGAU (.((.((((((((..((((((.....))))))..))))))............((((((((((((.(((....)))....----------------....)))))))))))).)))).).. ( -26.80) >DroSim_CAF1 11610 120 + 1 CUAGCACACUUUGUCCUUAAGUAUAUCUUGGGAUUAAAGUAUAUAUGUAUAUGUGACCUCUGUUGCGAAAUUUCGAUCCUUUCUUCCCUACGGGAUGAAGAUAGAGGUCAUCGUCUUGAU (.((.((((((((..((((((.....))))))..))))))............((((((((((((.(((....))).....(((.((((...)))).))))))))))))))).)))).).. ( -33.90) >DroEre_CAF1 19847 120 + 1 CUAGCACACGUUGUCCUUAAGUAUAUCUUGGGAUUAAAGUAUAUAUGUAUAUGUGACCUCUGUGGUGAAAUUUCGAUCCCAUUUUCCCUUCGGGAUAAAGACAAAGGUCAUCGUCUUGAU .......(((.((.((((..........((((((..(((((((((....))))).(((.....)))...))))..)))))).....((....)).........)))).)).)))...... ( -26.60) >DroYak_CAF1 10820 120 + 1 CUAGCACACGUUGUCCUUAAGUAUAUCUUGGGAUUAAAGUAUAUAUGUAUAUGUGACCUCUGUGGCGAAUUUUCGAUCCCUUUUUUCCUUCGGGAUGAAGAUAGAGGUCAUCGUCUUGAU (.((.((((...(((((.(((.....))))))))....))............(((((((((((..(((....)))....((((..(((....))).))))))))))))))).)))).).. ( -31.60) >consensus CUAGCACACUUUGUCCUUAAGUAUAUCUUGGGAUUAAAGUAUAUAUGUAUAUGUGACCUCUGUUGCGAAAUUUCGAUCCCUUCUUCCCUACGGGAUGAAGAUAGAGGUCAUCGUCUUGAU (.((.((((((((..((((((.....))))))..))))))............(((((((((((..(((....)))....((((.((((...)))).))))))))))))))).)))).).. (-25.86 = -27.02 + 1.16)



| Location | 15,927,925 – 15,928,030 |
|---|---|
| Length | 105 |
| Sequences | 5 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 87.29 |
| Mean single sequence MFE | -27.06 |
| Consensus MFE | -22.34 |
| Energy contribution | -23.10 |
| Covariance contribution | 0.76 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.89 |
| Structure conservation index | 0.83 |
| SVM decision value | 1.07 |
| SVM RNA-class probability | 0.910316 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15927925 105 + 22224390 AUAUAUGUAUAUGUGACCUCUGUUGCGAAAUUUCGAUCCCUUCUUCCCUACGGGAUGAAGAUAGAGGUCAUCGUCUUGAUGCGUAGAUUAU-AUAUAAGAUUGCGG ...((((((((.(((((((((((..(((....)))....((((.((((...)))).))))))))))))))).((((........)))))))-)))))......... ( -31.20) >DroSec_CAF1 26476 89 + 1 AUAUAUGUAUAUGUGACCUCUGUUGCGAAAUUUCGAUCC----------------UGAAGAUAGAGGUCAUCGUCUUGAUGCGUAGAUUAU-AUAUAAGAUUGGGG ...((((((((.((((((((((((.(((....)))....----------------....)))))))))))).((((........)))))))-)))))......... ( -22.90) >DroSim_CAF1 11650 105 + 1 AUAUAUGUAUAUGUGACCUCUGUUGCGAAAUUUCGAUCCUUUCUUCCCUACGGGAUGAAGAUAGAGGUCAUCGUCUUGAUGCGUAGAUUAU-AUAUAAGAUUGGGG ...((((((((.((((((((((((.(((....))).....(((.((((...)))).))))))))))))))).((((........)))))))-)))))......... ( -30.00) >DroEre_CAF1 19887 105 + 1 AUAUAUGUAUAUGUGACCUCUGUGGUGAAAUUUCGAUCCCAUUUUCCCUUCGGGAUAAAGACAAAGGUCAUCGUCUUGAUGCGUAGAUUAU-AUAUAAGAUUGUAG ...((((((((.(((((((.(((((.((........))))......((....))......))).))))))).((((........)))))))-)))))......... ( -23.20) >DroYak_CAF1 10860 106 + 1 AUAUAUGUAUAUGUGACCUCUGUGGCGAAUUUUCGAUCCCUUUUUUCCUUCGGGAUGAAGAUAGAGGUCAUCGUCUUGAUGCGUUGAUUAUUAUAUUAUAUUGGGG ............(((((((((((..(((....)))....((((..(((....))).)))))))))))))))...((..(((...((((......)))))))..)). ( -28.00) >consensus AUAUAUGUAUAUGUGACCUCUGUUGCGAAAUUUCGAUCCCUUCUUCCCUACGGGAUGAAGAUAGAGGUCAUCGUCUUGAUGCGUAGAUUAU_AUAUAAGAUUGGGG ...((((((((((((((((((((..(((....)))....((((.((((...)))).)))))))))))))).)))....)))))))..................... (-22.34 = -23.10 + 0.76)

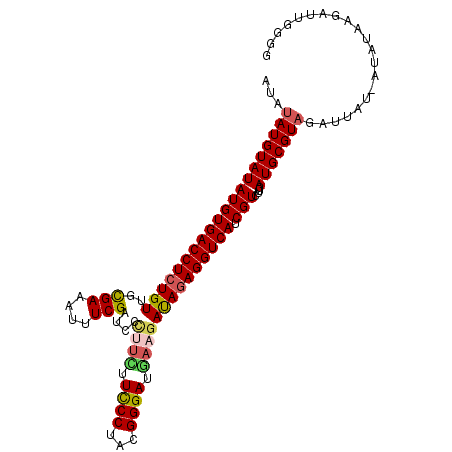
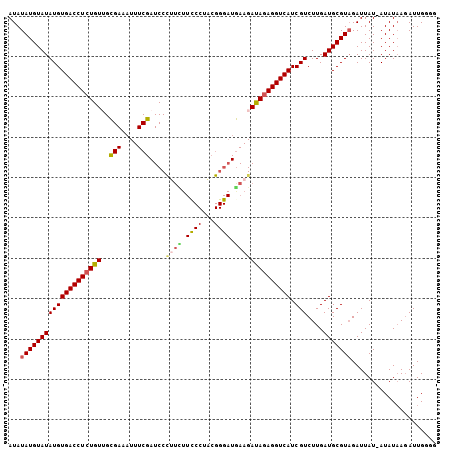
| Location | 15,927,925 – 15,928,030 |
|---|---|
| Length | 105 |
| Sequences | 5 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 87.29 |
| Mean single sequence MFE | -19.68 |
| Consensus MFE | -13.30 |
| Energy contribution | -14.78 |
| Covariance contribution | 1.48 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.94 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.07 |
| SVM RNA-class probability | 0.570993 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15927925 105 - 22224390 CCGCAAUCUUAUAU-AUAAUCUACGCAUCAAGACGAUGACCUCUAUCUUCAUCCCGUAGGGAAGAAGGGAUCGAAAUUUCGCAACAGAGGUCACAUAUACAUAUAU ..........((((-((..(((........))).(.((((((((.(((((.((((...)))).)))))...(((....)))....)))))))))......)))))) ( -23.80) >DroSec_CAF1 26476 89 - 1 CCCCAAUCUUAUAU-AUAAUCUACGCAUCAAGACGAUGACCUCUAUCUUCA----------------GGAUCGAAAUUUCGCAACAGAGGUCACAUAUACAUAUAU ..........((((-((..(((........))).(.(((((((((((....----------------.)))(((....)))....)))))))))......)))))) ( -13.60) >DroSim_CAF1 11650 105 - 1 CCCCAAUCUUAUAU-AUAAUCUACGCAUCAAGACGAUGACCUCUAUCUUCAUCCCGUAGGGAAGAAAGGAUCGAAAUUUCGCAACAGAGGUCACAUAUACAUAUAU ..........((((-((..(((........))).(.((((((((...(((.((((...)))).))).(...(((....)))...))))))))))......)))))) ( -20.50) >DroEre_CAF1 19887 105 - 1 CUACAAUCUUAUAU-AUAAUCUACGCAUCAAGACGAUGACCUUUGUCUUUAUCCCGAAGGGAAAAUGGGAUCGAAAUUUCACCACAGAGGUCACAUAUACAUAUAU ..........((((-((..(((........))).(.((((((((((.....((((...))))....((((........)).)))))))))))))......)))))) ( -21.50) >DroYak_CAF1 10860 106 - 1 CCCCAAUAUAAUAUAAUAAUCAACGCAUCAAGACGAUGACCUCUAUCUUCAUCCCGAAGGAAAAAAGGGAUCGAAAAUUCGCCACAGAGGUCACAUAUACAUAUAU .....(((((.((((........((........)).((((((((.(((((.....)))))......((((........)).))..))))))))..))))..))))) ( -19.00) >consensus CCCCAAUCUUAUAU_AUAAUCUACGCAUCAAGACGAUGACCUCUAUCUUCAUCCCGAAGGGAAAAAGGGAUCGAAAUUUCGCAACAGAGGUCACAUAUACAUAUAU ..................................(.((((((((.(((((.((((...)))).)))))((........)).....)))))))))............ (-13.30 = -14.78 + 1.48)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:09:03 2006